8WOS
 
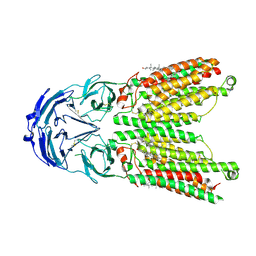 | | Cryo-EM structure of human SIDT1 protein with C1 symmetry at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
8WOT
 
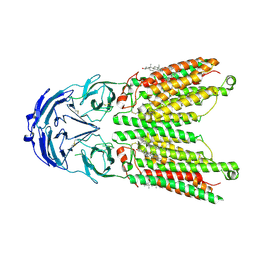 | | Cryo-EM structure of human SIDT1 protein with C2 symmetry at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
2H3D
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor in Complex with Nicotinamide Mononuleotide | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide phosphoribosyltransferase | | Authors: | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
8X3S
 
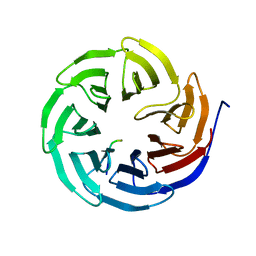 | | Crystal structure of human WDR5 in complex with PTEN | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, WD repeat-containing protein 5 | | Authors: | Liu, Y, Huang, X, Shang, X. | | Deposit date: | 2023-11-14 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The NTE domain of PTEN alpha / beta promotes cancer progression by interacting with WDR5 via its SSSRRSS motif.
Cell Death Dis, 15, 2024
|
|
7YL3
 
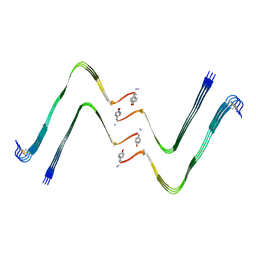 | | Structure of hIAPP-TF-type1 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Li, D, Zhang, X, Wang, Y, Zhu, P. | | Deposit date: | 2022-07-25 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A new polymorphism of human amylin fibrils with similar protofilaments and a conserved core.
Iscience, 25, 2022
|
|
7YL0
 
 | | Structure of hIAPP-TF-type2 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Li, D, Zhang, X, Wang, Y, Zhu, P. | | Deposit date: | 2022-07-25 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A new polymorphism of human amylin fibrils with similar protofilaments and a conserved core.
Iscience, 25, 2022
|
|
7YL7
 
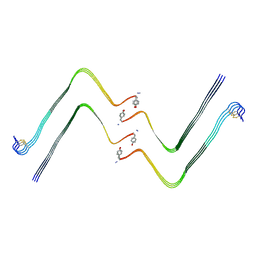 | | Structure of hIAPP-TF-type3 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Li, D, Zhang, X. | | Deposit date: | 2022-07-25 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A new polymorphism of human amylin fibrils with similar protofilaments and a conserved core.
Iscience, 25, 2022
|
|
7YKW
 
 | |
7FIL
 
 | |
7LCI
 
 | | PF 06882961 bound to the glucagon-like peptide-1 receptor (GLP-1R):Gs complex | | Descriptor: | 2-[(4-{6-[(4-cyano-2-fluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-{[(2S)-oxetan-2-yl]methyl}-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Johnson, R.M, Drulyte, I, Yu, L, Kotecha, A, Danev, R, Wootten, D, Zhang, X, Sexton, P.M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-01-20 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Evolving cryo-EM structural approaches for GPCR drug discovery.
Structure, 29, 2021
|
|
7LCJ
 
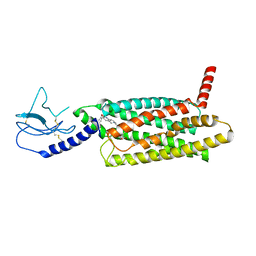 | | PF 06882961 bound to the glucagon-like peptide-1 receptor (GLP-1R):Gs complex | | Descriptor: | 2-[(4-{6-[(4-cyano-2-fluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-{[(2S)-oxetan-2-yl]methyl}-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor | | Authors: | Belousoff, M.J, Johnson, R.M, Drulyte, I, Yu, L, Kotecha, A, Danev, R, Wootten, D, Zhang, X, Sexton, P.M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-01-20 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Evolving cryo-EM structural approaches for GPCR drug discovery.
Structure, 29, 2021
|
|
1SQT
 
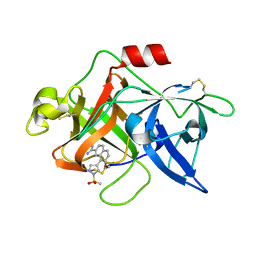 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 7-METHOXY-8-[1-(METHYLSULFONYL)-1H-PYRAZOL-4-YL]NAPHTHALENE-2-CARBOXIMIDAMIDE, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhang, X, Mantei, R, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2004-03-19 | | Release date: | 2004-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
7LCK
 
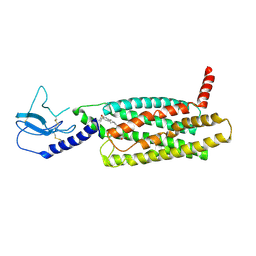 | | PF 06882961 bound to the glucagon-like peptide-1 receptor (GLP-1R) | | Descriptor: | 2-[(4-{6-[(4-cyano-2-fluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-{[(2S)-oxetan-2-yl]methyl}-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor | | Authors: | Belousoff, M.J, Johnson, R.M, Drulyte, I, Yu, L, Kotecha, A, Danev, R, Wootten, D, Zhang, X, Sexton, P.M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-01-20 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Evolving cryo-EM structural approaches for GPCR drug discovery.
Structure, 29, 2021
|
|
2H3B
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor 1 | | Descriptor: | Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
7FBI
 
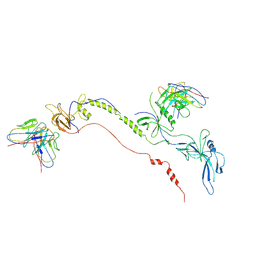 | | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5 | | Descriptor: | 3A3 heavy chain, 3A3 light chain, 3A5 heavy chain, ... | | Authors: | Zheng, Q, Li, S, Zha, Z, Hong, J, Chen, Y, Zhang, X. | | Deposit date: | 2021-07-10 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5
To Be Published
|
|
3TC1
 
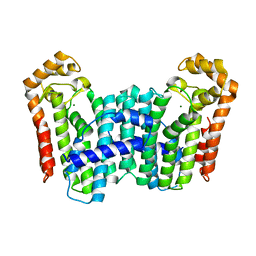 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Helicobacter pylori | | Descriptor: | MAGNESIUM ION, Octaprenyl Pyrophosphate Synthase | | Authors: | Zhang, J.Y, Zhang, X.L, Li, D.F, Zou, Q.M, Wang, D.C. | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Helicobacter pylori
To be Published
|
|
4M8N
 
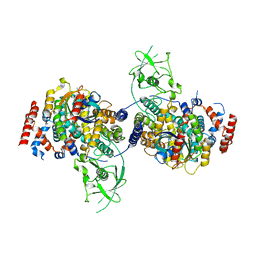 | | Crystal Structure of PlexinC1/Rap1B Complex | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pascoe, H.G, Wang, Y, Brautigam, C.A, He, H, Zhang, X. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Structural basis for activation and non-canonical catalysis of the Rap GTPase activating protein domain of plexin.
Elife, 2, 2013
|
|
7WB4
 
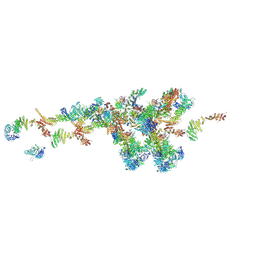 | | Cryo-EM structure of the NR subunit from X. laevis NPC | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Huang, G, Zhan, X, Shi, Y. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structure of the nuclear ring from Xenopus laevis nuclear pore complex.
Cell Res., 32, 2022
|
|
1LFC
 
 | | BOVINE LACTOFERRICIN (LFCINB), NMR, 20 STRUCTURES | | Descriptor: | LACTOFERRICIN | | Authors: | Hwang, P.M, Zhou, N, Shan, X, Arrowsmith, C.H, Vogel, H.J. | | Deposit date: | 1998-06-24 | | Release date: | 1998-11-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of lactoferricin B, an antimicrobial peptide derived from bovine lactoferrin.
Biochemistry, 37, 1998
|
|
7WKK
 
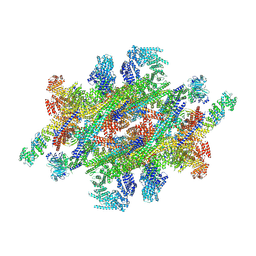 | | Cryo-EM structure of the IR subunit from X. laevis NPC | | Descriptor: | Aaas-prov protein, IL4I1 protein, MGC83295 protein, ... | | Authors: | Huang, G, Zhan, X, Shi, Y. | | Deposit date: | 2022-01-10 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the inner ring from the Xenopus laevis nuclear pore complex.
Cell Res., 32, 2022
|
|
7X2U
 
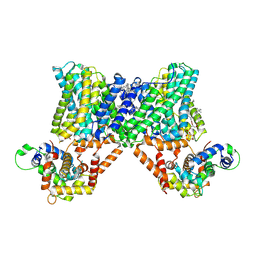 | | Structure of a human NHE3-CHP1 complex in the autoinhibited state | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Calcineurin B homologous protein 1, Sodium/hydrogen exchanger 3, ... | | Authors: | Dong, Y, Li, H, Gao, Y, Zhang, X.C, Zhao, Y. | | Deposit date: | 2022-02-26 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a human NHE3-CHP1 complex in the autoinhibited state
Sci Adv, 2022
|
|
3S8M
 
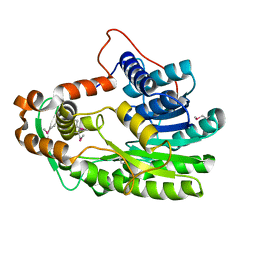 | | The Crystal Structure of FabV | | Descriptor: | Enoyl-ACP Reductase | | Authors: | Li, H, Zhang, X.L, Bi, L.J, He, J, Jiang, T. | | Deposit date: | 2011-05-29 | | Release date: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Determination of the Crystal Structure and Active Residues of FabV, the Enoyl-ACP Reductase from Xanthomonas oryzae.
Plos One, 6, 2011
|
|
7X5A
 
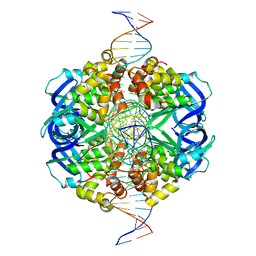 | | CryoEM structure of RuvA-Holliday junction complex | | Descriptor: | DNA (26-MER), Holliday junction ATP-dependent DNA helicase RuvA | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7Q
 
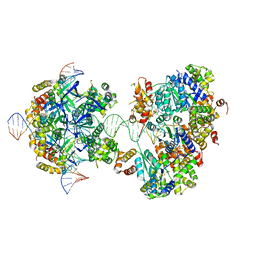 | | CryoEM structure of RuvA-RuvB-Holliday junction complex | | Descriptor: | DNA (26-MER), DNA (40-MER), Holliday junction ATP-dependent DNA helicase RuvA, ... | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7P
 
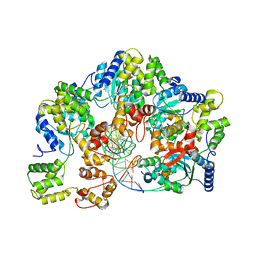 | | CryoEM structure of dsDNA-RuvB-RuvA domain3 complex | | Descriptor: | DNA, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
