4H42
 
 | | Synthesis of a Weak Basic uPA Inhibitor and Crystal Structure of Complex with uPA | | 分子名称: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, N-[(2-amino-1,3-benzothiazol-6-yl)carbonyl]glycine, Urokinase-type plasminogen activator | | 著者 | Yu, H.-Y, Gao, D, Zhang, X, Jiang, L.-G, Hong, Z.-B, Yuan, C, Fang, X, Wang, J.-D, Huang, M.-D. | | 登録日 | 2012-09-14 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Synthesis of a Weak Basic uPA Inhibitor and Crystal Structure of Complex with uPA
CHIN.J.STRUCT.CHEM., 32, 2013
|
|
4QNR
 
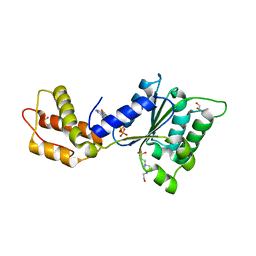 | | CRYSTAL STRUCTURE OF PSPF(1-265) E108Q MUTANT bound to ATP | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | 著者 | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | 登録日 | 2014-06-18 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.539 Å) | | 主引用文献 | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
4QOS
 
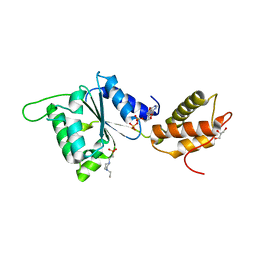 | | CRYSTAL STRUCTURE OF PSPF(1-265) E108Q MUTANT bound to ADP | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | 登録日 | 2014-06-20 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
4LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | 登録日 | 1991-01-25 | | 公開日 | 1992-07-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
6IWW
 
 | | Cryo-EM structure of the S. typhimurium oxaloacetate decarboxylase beta-gamma sub-complex | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Oxaloacetate decarboxylase beta chain, Probable oxaloacetate decarboxylase gamma chain | | 著者 | Xu, X, Shi, H, Zhang, X, Xiang, S. | | 登録日 | 2018-12-08 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insights into sodium transport by the oxaloacetate decarboxylase sodium pump.
Elife, 9, 2020
|
|
4QNM
 
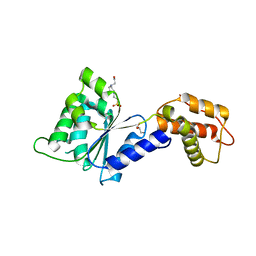 | | CRYSTAL STRUCTURE of PSPF(1-265) E108Q MUTANT | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Psp operon transcriptional activator | | 著者 | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | 登録日 | 2014-06-18 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.628 Å) | | 主引用文献 | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
1EPY
 
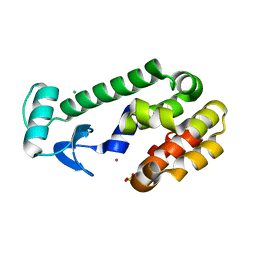 | | T4 LYSOZYME MUTANT, T21H/C54T/C97A/Q141H/T142H | | 分子名称: | CHLORIDE ION, COBALT (II) ION, LYSOZYME, ... | | 著者 | Wray, J.W, Baase, W.A, Ostheimer, G.J, Zhang, X.-J, Matthews, B.W. | | 登録日 | 2000-03-29 | | 公開日 | 2000-04-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
1F4H
 
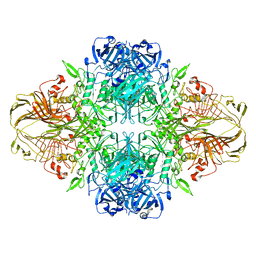 | | E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) | | 分子名称: | BETA-GALACTOSIDASE, MAGNESIUM ION | | 著者 | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | 登録日 | 2000-06-07 | | 公開日 | 2001-02-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1F4A
 
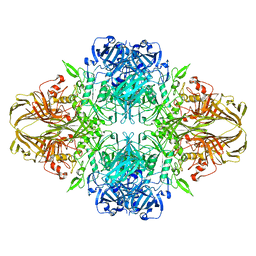 | | E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) | | 分子名称: | BETA-GALACTOSIDASE, MAGNESIUM ION | | 著者 | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | 登録日 | 2000-06-07 | | 公開日 | 2001-02-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1F6W
 
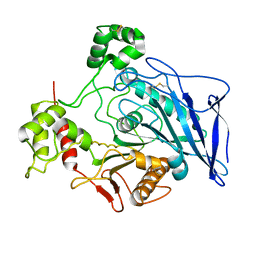 | |
1G55
 
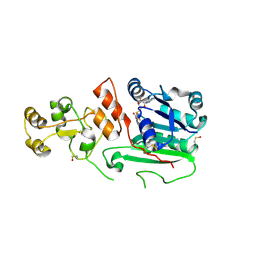 | | Structure of human DNMT2, an enigmatic DNA methyltransferase homologue | | 分子名称: | BETA-MERCAPTOETHANOL, DNA CYTOSINE METHYLTRANSFERASE DNMT2, GLYCEROL, ... | | 著者 | Dong, A, Yoder, J.A, Zhang, X, Zhou, L, Bestor, T.H, Cheng, X. | | 登録日 | 2000-10-30 | | 公開日 | 2001-01-17 | | 最終更新日 | 2018-06-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA.
Nucleic Acids Res., 29, 2001
|
|
4DFF
 
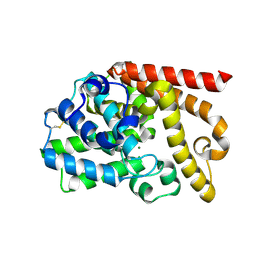 | | The SAR development of dihydroimidazoisoquinoline derivatives as phosphodiesterase 10A inhibitors for the treatment of schizophrenia | | 分子名称: | 8,9-dimethoxy-1-(1,3-thiazol-5-yl)-5,6-dihydroimidazo[5,1-a]isoquinoline, MAGNESIUM ION, ZINC ION, ... | | 著者 | Ho, G.D, Seganish, W.M, Bercovici, A, Tulshian, D, Greenlee, W.J, Van Rijn, R, Hruza, A, Xiao, L, Rindgen, D, Mullins, D, Guzzi, M, Zhang, X, Bleichardt, C, Hodgson, R. | | 登録日 | 2012-01-23 | | 公開日 | 2012-03-14 | | 最終更新日 | 2012-04-04 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | The SAR development of dihydroimidazoisoquinoline derivatives as phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4DHI
 
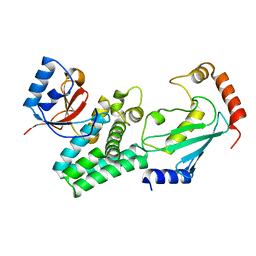 | | Structure of C. elegans OTUB1 bound to human UBC13 | | 分子名称: | Ubiquitin thioesterase otubain-like, Ubiquitin-conjugating enzyme E2 N | | 著者 | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | 登録日 | 2012-01-27 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
1BML
 
 | |
4N22
 
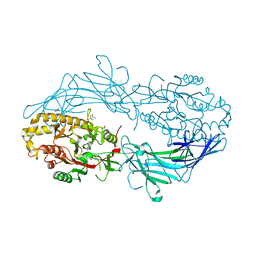 | | Crystal structure of Protein Arginine Deiminase 2 (50 uM Ca2+) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | 登録日 | 2013-10-04 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.889 Å) | | 主引用文献 | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N2I
 
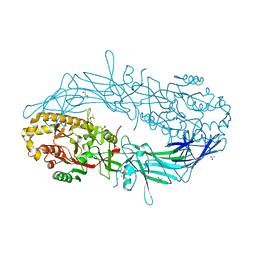 | | Crystal structure of Protein Arginine Deiminase 2 (D177A, 10 mM Ca2+) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | 登録日 | 2013-10-04 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N2D
 
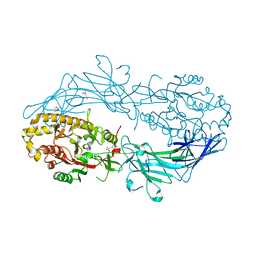 | | Crystal structure of Protein Arginine Deiminase 2 (D123N, 0 mM Ca2+) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Protein-arginine deiminase type-2 | | 著者 | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | 登録日 | 2013-10-04 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
7V8I
 
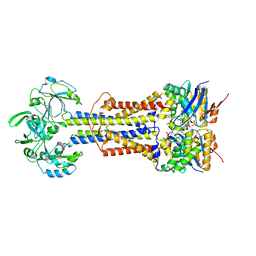 | | LolCD(E171Q)E with bound AMPPNP in nanodiscs | | 分子名称: | Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolC, Lipoprotein-releasing system transmembrane protein LolE, ... | | 著者 | Bei, W.W, Luo, Q.S, Shi, H.G, Zhang, X.Z, Huang, Y.H. | | 登録日 | 2021-08-23 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures of LolCDE reveal the molecular mechanism of bacterial lipoprotein sorting in Escherichia coli.
Plos Biol., 20, 2022
|
|
4N26
 
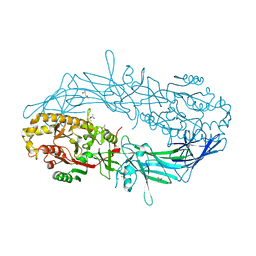 | | Crystal structure of Protein Arginine Deiminase 2 (500 uM Ca2+) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | 登録日 | 2013-10-04 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.943 Å) | | 主引用文献 | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
7V8M
 
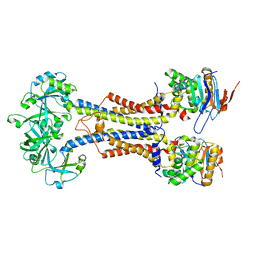 | | LolCDE-apo in nanodiscs | | 分子名称: | Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolC, Lipoprotein-releasing system transmembrane protein LolE | | 著者 | Luo, Q.S, Bei, W.W, Shi, H.G, Zhang, X.Z, Huang, Y.H. | | 登録日 | 2021-08-23 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-EM structures of LolCDE reveal the molecular mechanism of bacterial lipoprotein sorting in Escherichia coli.
Plos Biol., 20, 2022
|
|
4N2L
 
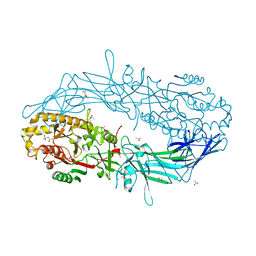 | | Crystal structure of Protein Arginine Deiminase 2 (Q350A, 10 mM Ca2+) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | 登録日 | 2013-10-05 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
7V8L
 
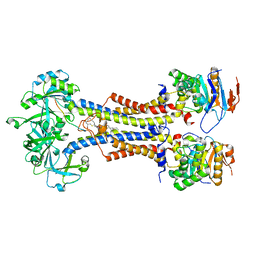 | | LolCDE with bound RcsF in nanodiscs | | 分子名称: | (2R)-3-{[(2S)-3-HYDROXY-2-(PALMITOYLAMINO)PROPYL]THIO}PROPANE-1,2-DIYL DIHEXADECANOATE, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolC, ... | | 著者 | Bei, W.W, Luo, Q.S, Shi, H.G, Zhang, X.Z, Huang, Y.H. | | 登録日 | 2021-08-23 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM structures of LolCDE reveal the molecular mechanism of bacterial lipoprotein sorting in Escherichia coli.
Plos Biol., 20, 2022
|
|
7W72
 
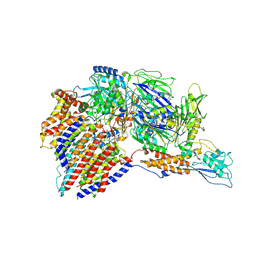 | | Structure of a human glycosylphosphatidylinositol (GPI) transamidase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GPI transamidase component PIG-S, ... | | 著者 | Zhang, H, Su, J, Li, B, Gao, Y, Zhang, X.C, Zhao, Y. | | 登録日 | 2021-12-02 | | 公開日 | 2022-02-16 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure of human glycosylphosphatidylinositol transamidase.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7WB4
 
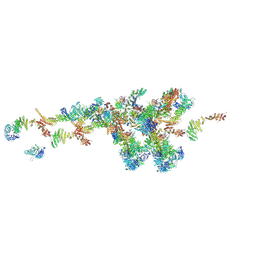 | | Cryo-EM structure of the NR subunit from X. laevis NPC | | 分子名称: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | 著者 | Huang, G, Zhan, X, Shi, Y. | | 登録日 | 2021-12-15 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (5.6 Å) | | 主引用文献 | Cryo-EM structure of the nuclear ring from Xenopus laevis nuclear pore complex.
Cell Res., 32, 2022
|
|
2LIT
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in reduced states | | 分子名称: | Cytochrome c iso-1, HEME C | | 著者 | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | 登録日 | 2011-08-31 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
