5C9D
 
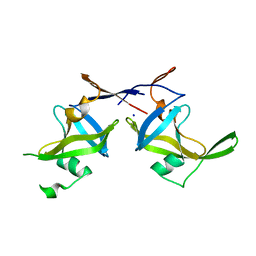 | | Crystal structure of a retropepsin-like aspartic protease from Rickettsia conorii | | Descriptor: | ApRick protease, SODIUM ION | | Authors: | Li, M, Gustchina, A, Cruz, R, Simoes, M, Curto, P, Martinez, J, Faro, C, Simoes, I, Wlodawer, A. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of RC1339/APRc from Rickettsia conorii, a retropepsin-like aspartic protease
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6M8W
 
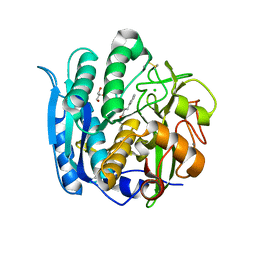 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR AIAF | | Descriptor: | AIAF PEPTIDE INHIBITOR, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-22 | | Release date: | 2018-10-24 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
5C9F
 
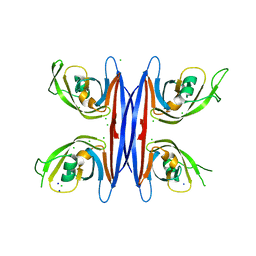 | | Crystal structure of a retropepsin-like aspartic protease from Rickettsia conorii | | Descriptor: | ApRick protease, CHLORIDE ION, SODIUM ION | | Authors: | Li, M, Gustchina, A, Cruz, R, Simoes, M, Curto, P, Martinez, J, Faro, C, Simoes, I, Wlodawer, A. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of RC1339/APRc from Rickettsia conorii, a retropepsin-like aspartic protease.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
6N2I
 
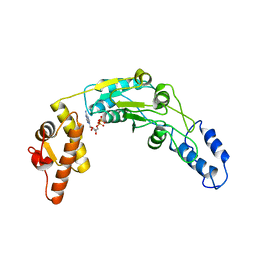 | | Lon protease AAA+ domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA-binding ATP-dependent protease La | | Authors: | Botos, I, Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2018-11-13 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | New insights into structural and functional relationships between LonA proteases and ClpB chaperones.
Febs Open Bio, 9, 2019
|
|
3SLZ
 
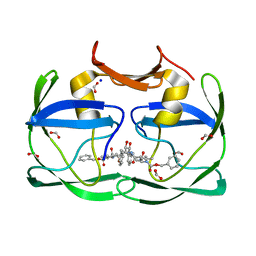 | | The crystal structure of XMRV protease complexed with TL-3 | | Descriptor: | FORMIC ACID, SODIUM ION, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate, ... | | Authors: | Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-06-27 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and biochemical characterization of the inhibitor complexes of xenotropic murine leukemia virus-related virus protease.
Febs J., 278, 2011
|
|
3SM2
 
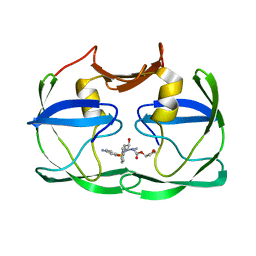 | | The crystal structure of XMRV protease complexed with Amprenavir | | Descriptor: | gag-pro-pol polyprotein, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-06-27 | | Release date: | 2011-10-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of the inhibitor complexes of xenotropic murine leukemia virus-related virus protease.
Febs J., 278, 2011
|
|
2ANE
 
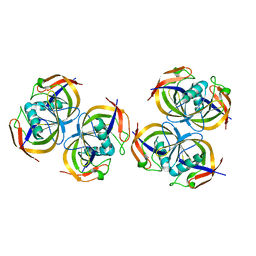 | | Crystal structure of N-terminal domain of E.Coli Lon Protease | | Descriptor: | ATP-dependent protease La | | Authors: | Li, M, Rasulova, F, Melnikov, E.E, Rotanova, T.V, Gustchina, A, Maurizi, M.R, Wlodawer, A. | | Deposit date: | 2005-08-11 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of the N-terminal domain of E. coli Lon protease.
Protein Sci., 14, 2005
|
|
2FMB
 
 | | EIAV PROTEASE COMPLEXED WITH AN INHIBITOR LP-130 | | Descriptor: | 4-[2-(2-ACETYLAMINO-3-NAPHTALEN-1-YL-PROPIONYLAMINO)-4-METHYL-PENTANOYLAMINO]-3-HYDROXY-6-METHYL-HEPTANOIC ACID [1-(1-CARBAMOYL-2-NAPHTHALEN-1-YL-ETHYLCARBAMOYL)-PROPYL]-AMIDE, EQUINE INFECTIOUS ANEMIA VIRUS PROTEASE | | Authors: | Kervinen, J, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-07-13 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a universal inhibitor of retroviral proteases: comparative analysis of the interactions of LP-130 complexed with proteases from HIV-1, FIV, and EIAV.
Protein Sci., 7, 1998
|
|
2B7F
 
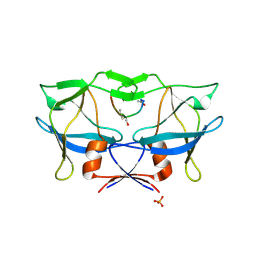 | | Crystal structure of human T-cell leukemia virus protease, a novel target for anti-cancer design | | Descriptor: | (ACE)APQV(STA)VMHP peptide, HTLV protease, PHOSPHATE ION | | Authors: | Li, M, Laco, G.S, Jaskolski, M, Rozycki, J, Alexandratos, J, Wlodawer, A, Gustchina, A. | | Deposit date: | 2005-10-04 | | Release date: | 2005-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human T cell leukemia virus protease, a novel target for anticancer drug design
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3FIV
 
 | | CRYSTAL STRUCTURE OF FELINE IMMUNODEFICIENCY VIRUS PROTEASE COMPLEXED WITH A SUBSTRATE | | Descriptor: | ACE-ALN-VAL-LEU-ALA-GLU-ALN-NH2, FELINE IMMUNODEFICIENCY VIRUS PROTEASE, SULFATE ION | | Authors: | Schalk-Hihi, C, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1997-07-09 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the inactive D30N mutant of feline immunodeficiency virus protease complexed with a substrate and an inhibitor.
Biochemistry, 36, 1997
|
|
3QRV
 
 | |
3QS1
 
 | | Crystal structure of KNI-10006 complex of Plasmepsin I (PMI) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Plasmepsin-1 | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-19 | | Release date: | 2011-05-11 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the free and inhibited forms of plasmepsin I (PMI) from Plasmodium falciparum.
J.Struct.Biol., 175, 2011
|
|
3SM1
 
 | |
3QVC
 
 | |
3QVI
 
 | | Crystal structure of KNI-10395 bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-{[S-methyl-N-(phenylacetyl)-L-cysteinyl]amino}-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-25 | | Release date: | 2011-10-12 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the activation and inhibition of histo-aspartic protease from Plasmodium falciparum.
Biochemistry, 50, 2011
|
|
3FNU
 
 | | Crystal structure of KNI-10006 bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, HAP protein | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2008-12-26 | | Release date: | 2009-05-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the histo-aspartic protease (HAP) from Plasmodium falciparum.
J.Mol.Biol., 388, 2009
|
|
1ZVK
 
 | | Structure of Double mutant, D164N, E78H of Kumamolisin-As | | Descriptor: | CALCIUM ION, kumamolisin-As | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Nakayama, T. | | Deposit date: | 2005-06-02 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Processing, catalytic activity and crystal structures of kumamolisin-As with an engineered active site.
Febs J., 273, 2006
|
|
1ZVJ
 
 | | Structure of Kumamolisin-AS mutant, D164N | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin-As | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Nakayama, T. | | Deposit date: | 2005-06-02 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Processing, catalytic activity and crystal structures of kumamolisin-As with an engineered active site.
Febs J., 273, 2006
|
|
2NR6
 
 | | Crystal structure of the complex of antibody and the allergen Bla g 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody heavy chain, ... | | Authors: | Li, M, Gustchina, A, Wlodawer, A, Pomes, A, Wunschmann, S. | | Deposit date: | 2006-11-01 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of a dimerized cockroach allergen Bla g 2 complexed with a monoclonal antibody.
J.Biol.Chem., 283, 2008
|
|
3FNS
 
 | |
2P3D
 
 | | Crystal Structure of the multi-drug resistant mutant subtype F HIV protease complexed with TL-3 inhibitor | | Descriptor: | Pol protein, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Sanches, M, Krauchenco, S, Martins, N.H, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Characterization of B and non-B Subtypes of HIV-Protease: Insights into the Natural Susceptibility to Drug Resistance Development.
J.Mol.Biol., 369, 2007
|
|
2P3A
 
 | | Crystal Structure of the multi-drug resistant mutant subtype B HIV protease complexed with TL-3 inhibitor | | Descriptor: | benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate, protease | | Authors: | Sanches, M, Krauchenco, S, Martins, N.H, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Characterization of B and non-B Subtypes of HIV-Protease: Insights into the Natural Susceptibility to Drug Resistance Development.
J.Mol.Biol., 369, 2007
|
|
1G0V
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT, MVV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEASE A INHIBITOR 3, PROTEINASE A, ... | | Authors: | Phylip, L.H, Lees, W, Brownsey, B.G, Bur, D, Dunn, B.M, Winther, J, Gustchina, A, Li, M, Copeland, T, Wlodawer, A, Kay, J. | | Deposit date: | 2000-10-09 | | Release date: | 2001-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The potency and specificity of the interaction between the IA3 inhibitor and its target aspartic proteinase from Saccharomyces cerevisiae.
J.Biol.Chem., 276, 2001
|
|
3FNT
 
 | | Crystal structure of pepstatin A bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, HAP protein, Inhibitor, ... | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2008-12-26 | | Release date: | 2009-05-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structures of the histo-aspartic protease (HAP) from plasmodium falciparum.
J.Mol.Biol., 388, 2009
|
|
3EMY
 
 | | Crystal structure of Trichoderma reesei aspartic proteinase complexed with pepstatin A | | Descriptor: | Pepstatin, Trichoderma reesei Aspartic protease | | Authors: | Nascimento, A.S, Krauchenco, S, Golubev, A.M, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-07 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Statistical coupling analysis of aspartic proteinases based on crystal
structures of the Trichoderma reesei enzyme and its complex with pepstatin A.
J.Mol.Biol., 382, 2008
|
|
