6WKS
 
 | | Structure of SARS-CoV-2 nsp16/nsp10 in complex with RNA cap analogue (m7GpppA) and S-adenosylmethionine | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, ADENOSINE, ... | | Authors: | Gupta, Y.K, Viswanathan, T, Arya, S, Qi, S, Misra, A, Chan, S.-H. | | Deposit date: | 2020-04-16 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of RNA cap modification by SARS-CoV-2.
Nat Commun, 11, 2020
|
|
4ZCF
 
 | | Structural basis of asymmetric DNA methylation and ATP-triggered long-range diffusion by EcoP15I | | Descriptor: | ADENOSINE MONOPHOSPHATE, CALCIUM ION, DNA 20-mer AATCATAGTCTACTGCTGTA, ... | | Authors: | Gupta, Y.K, Chan, S.H, Xu, S.Y, Aggarwal, A.K. | | Deposit date: | 2015-04-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of asymmetric DNA methylation and ATP-triggered long-range diffusion by EcoP15I.
Nat Commun, 6, 2015
|
|
3BSB
 
 | | Crystal Structure of Human Pumilio1 in Complex with CyclinB reverse RNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*AP*UP*GP*UP*U)-3', Pumilio homolog 1 | | Authors: | Gupta, Y.K, Nair, D.T, Wharton, R.P, Aggarwal, A.K. | | Deposit date: | 2007-12-23 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity.
Structure, 16, 2008
|
|
3BSX
 
 | | Crystal Structure of Human Pumilio 1 in complex with Puf5 RNA | | Descriptor: | 5'-R(*UP*UP*GP*UP*AP*AP*UP*AP*UP*UP*A)-3', Pumilio homolog 1 | | Authors: | Gupta, Y.K, Nair, D.T, Wharton, R.P, Aggarwal, A.K. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity.
Structure, 16, 2008
|
|
7LW3
 
 | | Structure of SARS-CoV-2 nsp16/nsp10 complex in presence of Cap-1 analog (m7GpppAmU) and SAH | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Gupta, Y.K, Viswanathan, T, Misra, A, Qi, S. | | Deposit date: | 2021-02-27 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A metal ion orients SARS-CoV-2 mRNA to ensure accurate 2'-O methylation of its first nucleotide.
Nat Commun, 12, 2021
|
|
7LW4
 
 | | Structure of SARS-CoV-2 nsp16/nsp10 complex in presence of S-adenosyl-L-homocysteine (SAH) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Gupta, Y.K, Viswanathan, T, Misra, A, Qi, S. | | Deposit date: | 2021-02-27 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A metal ion orients SARS-CoV-2 mRNA to ensure accurate 2'-O methylation of its first nucleotide.
Nat Commun, 12, 2021
|
|
5L2X
 
 | | Crystal structure of human PrimPol ternary complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*GP*GP*TP*AP*GP*CP*(DDG))-3'), ... | | Authors: | Rechkoblit, O, Gupta, Y.K, Malik, R, Rajashankar, K.R, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of human PrimPol, a DNA polymerase with primase activity.
Sci Adv, 2, 2016
|
|
5HR4
 
 | | Structure of Type IIL restriction-modification enzyme MmeI in complex with DNA has implications for engineering of new specificities | | Descriptor: | CALCIUM ION, DNA (5'-D(P*GP*TP*TP*AP*TP*GP*TP*CP*GP*GP*AP*TP*A)-3'), DNA (5'-D(P*TP*AP*TP*CP*CP*GP*AP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Callahan, S.J, Luyten, Y.A, Gupta, Y.K, Wilson, G.G, Roberts, R.J, Morgan, R.D, Aggarwal, A.K. | | Deposit date: | 2016-01-22 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5964 Å) | | Cite: | Structure of Type IIL Restriction-Modification Enzyme MmeI in Complex with DNA Has Implications for Engineering New Specificities.
Plos Biol., 14, 2016
|
|
1RK9
 
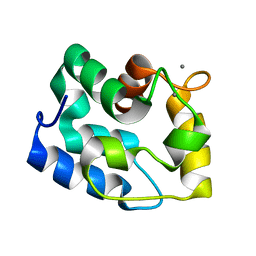 | | Solution Structure of Human alpha-Parvalbumin (Minimized Average Structure) | | Descriptor: | CALCIUM ION, Parvalbumin alpha | | Authors: | Baig, I, Bertini, I, Del Bianco, C, Gupta, Y.K, Lee, Y.-M, Luchinat, C, Quattrone, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-11-21 | | Release date: | 2004-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-based refinement strategy for the solution structure of human alpha-parvalbumin
Biochemistry, 43, 2004
|
|
1RJV
 
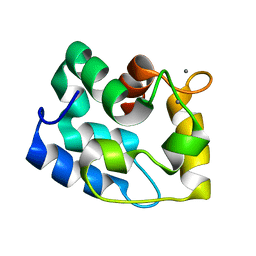 | | Solution Structure of Human alpha-Parvalbumin refined with a paramagnetism-based strategy | | Descriptor: | CALCIUM ION, Parvalbumin alpha | | Authors: | Baig, I, Bertini, I, Del Bianco, C, Gupta, Y.K, Lee, Y.M, Luchinat, C, Quattrone, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-11-20 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-Based Refinement Strategy for the Solution Structure of Human alpha-Parvalbumin.
Biochemistry, 43, 2004
|
|
