7L0R
 
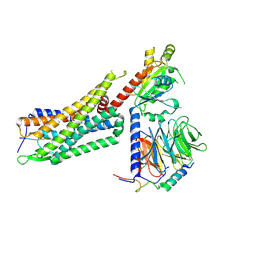 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0P
 
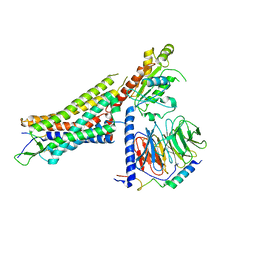 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0S
 
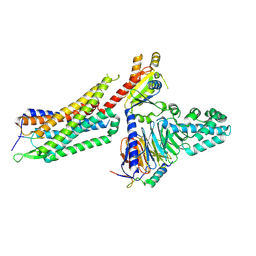 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0Q
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2BZA
 
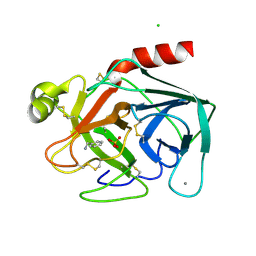 | | BOVINE PANCREAS BETA-TRYPSIN IN COMPLEX WITH BENZYLAMINE | | Descriptor: | BENZYLAMINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ota, N, Stroupe, C, Ferreira-Da-Silva, J.M.S, Shah, S.S, Mares-Guia, M, Brunger, A.T. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Non-Boltzmann thermodynamic integration (NBTI) for macromolecular systems: relative free energy of binding of trypsin to benzamidine and benzylamine.
Proteins, 37, 1999
|
|
4I45
 
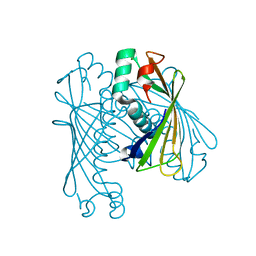 | | Crystal Structure of Orf6 protein from Photobacterium profundum, Mg2+-bound form | | Descriptor: | MAGNESIUM ION, ORF6 thioesterase | | Authors: | Rodriguez-Guilbe, M.M, Baerga-Ortiz, A, Schreiter, E.R. | | Deposit date: | 2012-11-27 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, Activity, and Substrate Selectivity of the Orf6 Thioesterase from Photobacterium profundum.
J.Biol.Chem., 288, 2013
|
|
1CE5
 
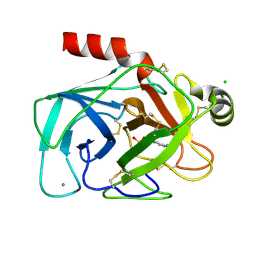 | | BOVINE PANCREAS BETA-TRYPSIN IN COMPLEX WITH BENZAMIDINE | | Descriptor: | BENZAMIDINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ota, N, Stroupe, C, Ferreira-Da-Silva, J.M.S, Shah, S.S, Mares-Guia, M, Brunger, A.T. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Non-Boltzmann thermodynamic integration (NBTI) for macromolecular systems: relative free energy of binding of trypsin to benzamidine and benzylamine.
Proteins, 37, 1999
|
|
2L2G
 
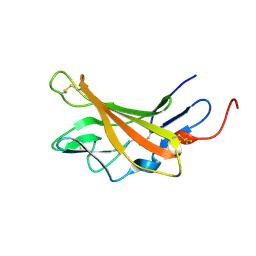 | | Solution structure of Opossum Domain 11 | | Descriptor: | IGF2R DOMAIN 11 | | Authors: | Williams, C, Hoppe, H, Rezgui, D, Rezgui, M, Frago, S, Ellis, R.Z, Wattana-Amorn, P, Prince, S.N, Zaccheo, O.J, Forbes, B, Jones, E.Y, Crump, M.P, Bassim, A.H. | | Deposit date: | 2010-08-18 | | Release date: | 2012-02-15 | | Last modified: | 2015-02-25 | | Method: | SOLUTION NMR | | Cite: | An exon splice enhancer primes IGF2:IGF2R binding site structure and function evolution.
Science, 338, 2012
|
|
5JRW
 
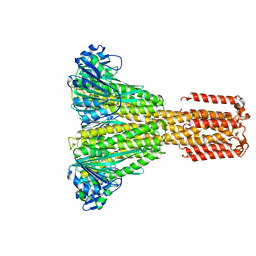 | |
5JTG
 
 | |
1NY3
 
 | | Crystal structure of ADP bound to MAP KAP kinase 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAP kinase-activated protein kinase 2 | | Authors: | Underwood, K.W, Parris, K.D, Federico, E, Mosyak, L, Shane, T, Taylor, M, Svenson, K, Liu, Y, Hsiao, C.L, Wolfrom, S, Maguire, M, Malakian, K, Telliez, J.B, Lin, L.L, Kriz, R.W, Seehra, J, Somers, W.S, Stahl, M.L. | | Deposit date: | 2003-02-11 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Catalytically active MAP KAP kinase 2 structures in complex with staurosporine and ADP reveal differences with the autoinhibited enzyme
Structure, 11, 2003
|
|
2AHE
 
 | | Crystal structure of a soluble form of CLIC4. intercellular chloride ion channel | | Descriptor: | Chloride intracellular channel protein 4 | | Authors: | Littler, D.R, Assaad, N.N, Harrop, S.J, Brown, L.J, Pankhurst, G.J, Luciani, P, Aguilar, M.-I, Mazzanti, M, Berryman, M.A, Breit, S.N, Curmi, P.M.G. | | Deposit date: | 2005-07-28 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the soluble form of the redox-regulated chloride ion channel protein CLIC4.
FEBS J., 272, 2005
|
|
4IPM
 
 | | Crystal structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with thiocellobiose | | Descriptor: | ACETATE ION, CALCIUM ION, GH7 family protein, ... | | Authors: | McGeehan, J.E, Martin, R.N.A, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2013-01-10 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HAQ
 
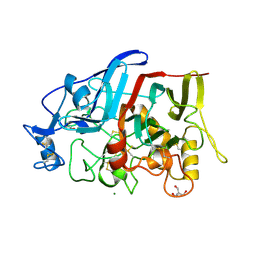 | | Crystal Structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with cellobiose and cellotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GH7 family protein, ... | | Authors: | Martin, R.N.A, McGeehan, J.E, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-27 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GWA
 
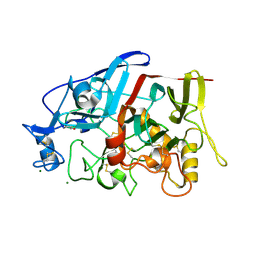 | | Crystal Structure of a GH7 Family Cellobiohydrolase from Limnoria quadripunctata | | Descriptor: | GH7 family protein, MAGNESIUM ION | | Authors: | McGeehan, J.E, Martin, R.N.A, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-01 | | Release date: | 2013-06-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HAP
 
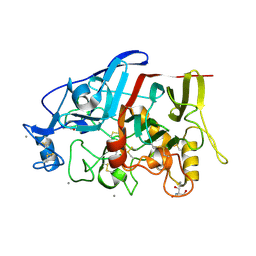 | | Crystal Structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with cellobiose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GH7 family protein, ... | | Authors: | Martin, R.N.A, McGeehan, J.E, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-27 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2NDA
 
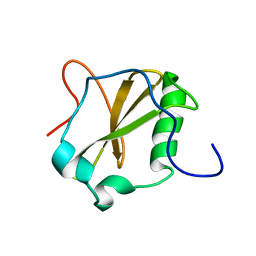 | | Solution structure of MapZ extracellular domain second subdomain | | Descriptor: | Mid-cell-anchored protein Z | | Authors: | Jean, N.L, Manuse, S, Guinot, M, Bougault, C.M, Grangeasse, C, Simorre, J.-P. | | Deposit date: | 2016-05-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the extracellular domain of the pneumococcal cell division site positioning protein MapZ.
Nat Commun, 7, 2016
|
|
2WCG
 
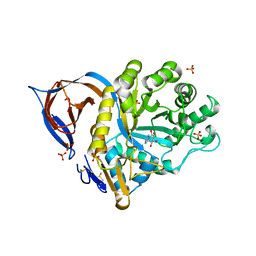 | | X-ray structure of acid-beta-glucosidase with N-octyl(cyclic guanidine)-nojirimycin in the active site | | Descriptor: | CHLORIDE ION, GLUCOSYLCERAMIDASE, N-[(3E,5R,6R,7S,8R,8AR)-5,6,7,8-TETRAHYDROXYHEXAHYDROIMIDAZO[1,5-A]PYRIDIN-3(2H)-YLIDENE]OCTAN-1-AMINIUM, ... | | Authors: | Brumshtein, B, Aguilar, M, Garcia-Moreno, M.I, Mellet, C.O, Garcia-Fernandez, J.M, Silman, I, Shaaltiel, Y, Aviezer, D, Sussman, J.L, Futerman, A.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 6-Amino-6-Deoxy-5,6-Di-N-(N'-Octyliminomethylidene)Nojirimycin: Synthesis, Biological Evaluation, and Crystal Structure in Complex with Acid Beta-Glucosidase.
Chembiochem, 10, 2009
|
|
2ND9
 
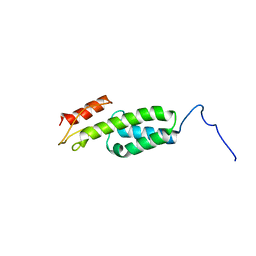 | | Solution structure of MapZ extracellular domain first subdomain | | Descriptor: | Mid-cell-anchored protein Z | | Authors: | Jean, N.L, Manuse, S, Guinot, M, Bougault, C.M, Grangeasse, C, Simorre, J.-P. | | Deposit date: | 2016-05-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the extracellular domain of the pneumococcal cell division site positioning protein MapZ.
Nat Commun, 7, 2016
|
|
1JDE
 
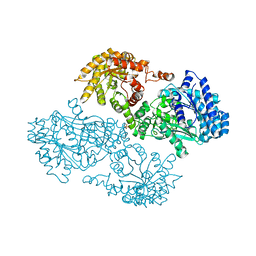 | | K22A mutant of pyruvate, phosphate dikinase | | Descriptor: | PYRUVATE, PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Ye, D, Wei, M, McGuire, M, Huang, K, Kapadia, G, Herzberg, O, Martin, B.M, Dunaway-Mariano, D. | | Deposit date: | 2001-06-13 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Investigation of the catalytic site within the ATP-grasp domain of Clostridium symbiosum pyruvate phosphate dikinase.
J.Biol.Chem., 276, 2001
|
|
2BBJ
 
 | | Crystal structure of the CorA Mg2+ transporter | | Descriptor: | divalent cation transport-related protein | | Authors: | Lunin, V.V, Dobrovetsky, E, Khutoreskaya, G, Zhang, R, Joachimiak, A, Bochkarev, A, Maguire, M.E, Edwards, A.M, Koth, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-10-17 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the CorA Mg2+ transporter
Nature, 440, 2006
|
|
2BBH
 
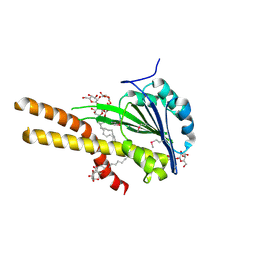 | | X-ray structure of T.maritima CorA soluble domain | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Lunin, V.V, Dobrovetsky, E, Khutoreskaya, G, Bochkarev, A, Maguire, M.E, Edwards, A.M, Koth, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-10-17 | | Release date: | 2005-12-13 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the CorA Mg2+ transporter.
Nature, 440, 2006
|
|
1TZ4
 
 | | [hPP19-23]-pNPY bound to DPC Micelles | | Descriptor: | neuropeptide Y,Pancreatic prohormone,neuropeptide Y | | Authors: | Lerch, M, Kamimori, H, Folkers, G, Aguilar, M.I, Beck-Sickinger, A.G, Zerbe, O. | | Deposit date: | 2004-07-09 | | Release date: | 2005-07-05 | | Last modified: | 2020-01-01 | | Method: | SOLUTION NMR | | Cite: | Strongly Altered Receptor Binding Properties in PP and NPY Chimeras Are Accompanied by Changes
in Structure and Membrane Binding
Biochemistry, 44, 2005
|
|
1TZ5
 
 | | [pNPY19-23]-hPP bound to DPC Micelles | | Descriptor: | Pancreatic prohormone,neuropeptide Y,Pancreatic prohormone | | Authors: | Lerch, M, Kamimori, H, Folkers, G, Aguilar, M.I, Beck-Sickinger, A.G, Zerbe, O. | | Deposit date: | 2004-07-09 | | Release date: | 2005-07-05 | | Last modified: | 2020-01-01 | | Method: | SOLUTION NMR | | Cite: | Strongly Altered Receptor Binding Properties in PP and NPY Chimeras Are Accompanied by Changes
in Structure and Membrane Binding
Biochemistry, 44, 2005
|
|
1S9Y
 
 | | Crystal Structure Analysis of NY-ESO-1 epitope analogue, SLLMWITQS, in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | Deposit date: | 2004-02-05 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
