2DGL
 
 | | Crystal structure of Escherichia coli GadB in complex with bromide | | Descriptor: | ACETIC ACID, BROMIDE ION, Glutamate decarboxylase beta, ... | | Authors: | Gruetter, M.G, Capitani, G, Gut, H. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
2DGM
 
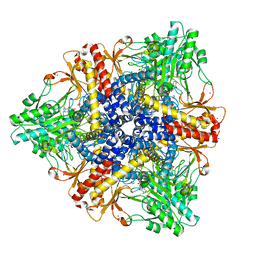 | | Crystal structure of Escherichia coli GadB in complex with iodide | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Gruetter, M.G, Capitani, G, Gut, H. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
2DGK
 
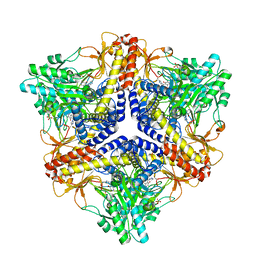 | | Crystal structure of an N-terminal deletion mutant of Escherichia coli GadB in an autoinhibited state (aldamine) | | Descriptor: | 1,2-ETHANEDIOL, Glutamate decarboxylase beta, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Gruetter, M.G, Capitani, G, Gut, H. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
1SBN
 
 | |
1RNE
 
 | |
1SIB
 
 | |
2FBE
 
 | | Crystal Structure of the PRYSPRY-domain | | Descriptor: | PREDICTED: similar to ret finger protein-like 1 | | Authors: | Gruetter, C, Briand, C, Capitani, G, Mittl, P.R, Gruetter, M.G. | | Deposit date: | 2005-12-09 | | Release date: | 2006-01-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the PRYSPRY-domain: Implications for autoinflammatory diseases
Febs Lett., 580, 2006
|
|
1HII
 
 | |
1HIH
 
 | |
3EO0
 
 | |
3EO1
 
 | |
2AXI
 
 | | HDM2 in complex with a beta-hairpin | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, SULFATE ION, Ubiquitin-protein ligase E3 Mdm2, ... | | Authors: | Mittl, P.R.E, Fasan, R, Robinson, J, Gruetter, M.G. | | Deposit date: | 2005-09-05 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Studies in a Family of beta-Hairpin Protein Epitope Mimetic Inhibitors of the p53-HDM2 Protein-Protein Interaction.
Chembiochem, 7, 2006
|
|
4CDL
 
 | | Crystal Structure of Retro-aldolase RA110.4-6 Complexed with Inhibitor 1-(6-methoxy-2-naphthalenyl)-1,3-butanedione | | Descriptor: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, STEROID DELTA-ISOMERASE | | Authors: | Pinkas, D.M, Studer, S, Obexer, R, Giger, L, Gruetter, M.G, Baker, D, Hilvert, D. | | Deposit date: | 2013-11-01 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active Site Plasticity of a Computationally Designed Retro-Aldolase Enzyme
To be Published
|
|
4BS0
 
 | | Crystal Structure of Kemp Eliminase HG3.17 E47N,N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, KEMP ELIMINASE HG3.17, SULFATE ION | | Authors: | Blomberg, R, Kries, H, Pinkas, D.M, Mittl, P.R.E, Gruetter, M.G, Privett, H.K, Mayo, S, Hilvert, D. | | Deposit date: | 2013-06-06 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Precision is Essential for Efficient Catalysis in an Evolved Kemp Eliminase
Nature, 503, 2013
|
|
4CG4
 
 | | Crystal structure of the CHS-B30.2 domains of TRIM20 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-ETHANETHIOL, PYRIN, ... | | Authors: | Weinert, C, Morger, D, Djekic, A, Mittl, P.R.E, Gruetter, M.G. | | Deposit date: | 2013-11-20 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Trim20 C-Terminal Coiled-Coil/B30.2 Fragment: Implications for the Recognition of Higher Order Oligomers
Sci.Rep., 5, 2015
|
|
2I1B
 
 | |
3NDV
 
 | | Crystal structure of the N-terminal beta-aminopeptidase BapA in complex with ampicillin | | Descriptor: | (2S,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, Beta-peptidyl aminopeptidase, ... | | Authors: | Merz, T, Heck, T, Geueke, B, Kohler, H.-P.E, Gruetter, M.G. | | Deposit date: | 2010-06-08 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures and inhibition of the beta-aminopeptidase BapA, a new ampicillin-recognizing member of the N-terminal nucleophile hydrolase family
To be Published
|
|
3NFB
 
 | | Crystal structure of the N-terminal beta-aminopeptidase BapA in complex with hydrolyzed ampicillin | | Descriptor: | (2S,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-peptidyl aminopeptidase, GLYCEROL, ... | | Authors: | Merz, T, Heck, T, Geueke, B, Kohler, H.-P.E, Gruetter, M.G. | | Deposit date: | 2010-06-10 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and inhibition of the beta-aminopeptidase BapA, a new ampicillin-recognizing member of the N-terminal nucleophile hydrolase family
To be Published
|
|
2MIB
 
 | |
2K7Z
 
 | |
2P2C
 
 | | Inhibition of caspase-2 by a designed ankyrin repeat protein (DARPin) | | Descriptor: | Caspase-2 | | Authors: | Roschitzki Voser, H, Briand, C, Capitani, G, Gruetter, M.G. | | Deposit date: | 2007-03-07 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Inhibition of Caspase-2 by a Designed Ankyrin Repeat Protein: Specificity, Structure, and Inhibition Mechanism.
Structure, 15, 2007
|
|
4J7W
 
 | | E3_5 DARPin L86A mutant | | Descriptor: | DARPin_E3_5_L86A | | Authors: | Seeger, M.A, Gruetter, M.G. | | Deposit date: | 2013-02-14 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, construction, and characterization of a second-generation DARPin library with reduced hydrophobicity.
Protein Sci., 22, 2013
|
|
4J8Y
 
 | | E3_5 DARPin D77S mutant | | Descriptor: | DARPin_E3_5_D77S | | Authors: | Seeger, M.A, Gruetter, M.G. | | Deposit date: | 2013-02-15 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, construction, and characterization of a second-generation DARPin library with reduced hydrophobicity.
Protein Sci., 22, 2013
|
|
4JB8
 
 | | Caspase-7 in Complex with DARPin C7_16 | | Descriptor: | Caspase-7 subunit p11, Caspase-7 subunit p20, DARPin C7_16 | | Authors: | Fluetsch, A, Seeger, M.A, Gruetter, M.G. | | Deposit date: | 2013-02-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, construction, and characterization of a second-generation DARPin library with reduced hydrophobicity.
Protein Sci., 22, 2013
|
|
4LSZ
 
 | | Caspase-7 in Complex with DARPin D7.18 | | Descriptor: | Caspase-7 subunit p10, Caspase-7 subunit p20, DARPin D7.18 | | Authors: | Fluetsch, A, Lukarska, M, Gruetter, M.G. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Combined inhibition of caspase 3 and caspase 7 by two highly selective DARPins slows down cellular demise.
Biochem.J., 461, 2014
|
|
