1RF8
 
 | | Solution structure of the yeast translation initiation factor eIF4E in complex with m7GDP and eIF4GI residues 393 to 490 | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic initiation factor 4F subunit p150, Eukaryotic translation initiation factor 4E, ... | | Authors: | Gross, J.D, Moerke, N.J, von der Haar, T, Lugovskoy, A.A, Sachs, A.B, McCarthy, J.E.G, Wagner, G. | | Deposit date: | 2003-11-07 | | Release date: | 2003-12-23 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E.
Cell(Cambridge,Mass.), 115, 2003
|
|
3OEO
 
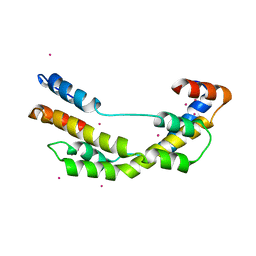 | | The crystal structure E. coli Spy | | Descriptor: | CADMIUM ION, Spheroplast protein Y | | Authors: | Kwon, E, Kim, D.Y, Gross, C.A, Gross, J.D, Kim, K.K. | | Deposit date: | 2010-08-13 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure Escherichia coli Spy.
Protein Sci., 19, 2010
|
|
2KEM
 
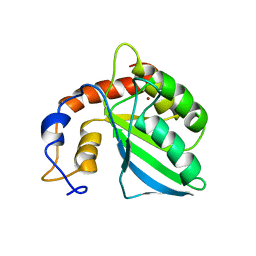 | | Extended structure of citidine deaminase domain of APOBEC3G | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Harjes, E, Gross, P.J, Chen, K, Lu, Y, Shindo, K, Nowarski, R, Gross, J.D, Kotler, M, Harris, R.S, Matsuo, H. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model
J.Mol.Biol., 389, 2009
|
|
6AM0
 
 | |
5KQ4
 
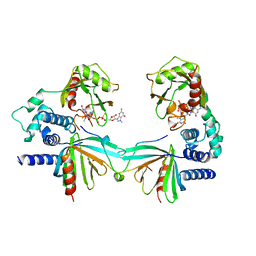 | | Crystal structure of S. pombe Dcp1/Dcp2 in complex with H. sapiens PNRC2 and synthetic cap analog | | Descriptor: | Proline-rich nuclear receptor coactivator 2, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-3~{H}-purin-7-ium-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-sulfanyl-phosphoryl] [[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-3~{H}-purin-7-ium-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-sulfanyl-phosphoryl]oxy-oxidanyl-phosphoryl] hydrogen phosphate, mRNA decapping complex subunit 2, ... | | Authors: | Mugridge, J.S, Ziemniak, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2016-07-05 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis of mRNA-cap recognition by Dcp1-Dcp2.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5KQ1
 
 | | Crystal structure of S. pombe Dcp1/Dcp2 in complex with H. sapiens PNRC2 | | Descriptor: | Proline-rich nuclear receptor coactivator 2, mRNA decapping complex subunit 2, mRNA-decapping enzyme subunit 1 | | Authors: | Mugridge, J.S, Ziemniak, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2016-07-05 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Structural basis of mRNA-cap recognition by Dcp1-Dcp2.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6P59
 
 | | Crystal structure of SIVrcm Vif-CBFbeta-ELOB-ELOC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Core-binding factor subunit beta, Elongin-B, ... | | Authors: | Binning, J.M, Chesarino, N.M, Emerman, M, Gross, J.D. | | Deposit date: | 2019-05-29 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.942214 Å) | | Cite: | Structural Basis for a Species-Specific Determinant of an SIV Vif Protein toward Hominid APOBEC3G Antagonism.
Cell Host Microbe, 26, 2019
|
|
3V67
 
 | | Periplasmic domain of Vibrio parahaemolyticus CpxA | | Descriptor: | Sensor protein CpxA | | Authors: | Kwon, E, Kim, D.Y, Ngo, T.D, Gross, J.D, Kim, K.K. | | Deposit date: | 2011-12-19 | | Release date: | 2012-09-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the periplasmic domain of Vibrio parahaemolyticus CpxA
Protein Sci., 21, 2012
|
|
1ZGW
 
 | | NMR structure of E. Coli Ada protein in complex with DNA | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*TP*TP*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*A)-3', 5'-D(*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*TP*AP*AP*TP*TP*TP*GP*C)-3', Ada polyprotein, ... | | Authors: | He, C, Hus, J.C, Sun, L.J, Zhou, P, Norman, D.P, Doetsch, V, Wei, H, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2005-04-22 | | Release date: | 2005-10-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A Methylation-Dependent Electrostatic Switch Controls DNA Repair and Transcriptional Activation by E. coli Ada.
Mol.Cell, 20, 2005
|
|
1TVC
 
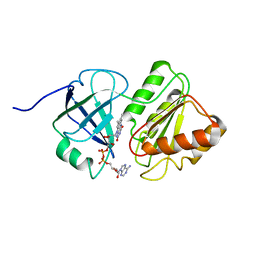 | | FAD and NADH binding domain of methane monooxygenase reductase from Methylococcus capsulatus (Bath) | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, METHANE MONOOXYGENASE COMPONENT C | | Authors: | Chatwood, L.L, Mueller, J, Gross, J.D, Wagner, G, Lippard, S.J. | | Deposit date: | 2004-06-29 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Flavin Domain from Soluble Methane Monooxygenase Reductase from Methylococcus capsulatus (Bath)
Biochemistry, 43, 2004
|
|
1U8B
 
 | | Crystal structure of the methylated N-ADA/DNA complex | | Descriptor: | 5'-D(*AP*AP*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*T)-3', 5'-D(*TP*AP*AP*AP*TP*T)-3', 5'-D(P*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*AP*T)-3', ... | | Authors: | He, C, Hus, J.-C, Sun, L.J, Zhou, P, Norman, D.P.G, Dotsch, V, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2004-08-05 | | Release date: | 2005-10-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A methylation-dependent electrostatic switch controls DNA repair and transcriptional activation by E. coli ada.
Mol.Cell, 20, 2005
|
|
4KG4
 
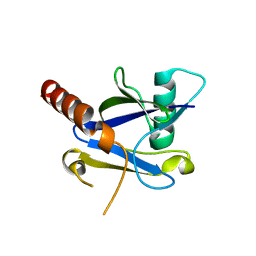 | |
4KG3
 
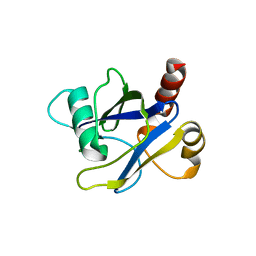 | |
4K6E
 
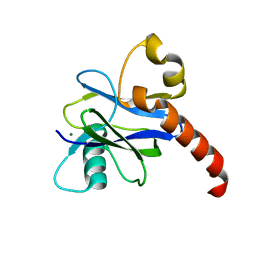 | |
8CX2
 
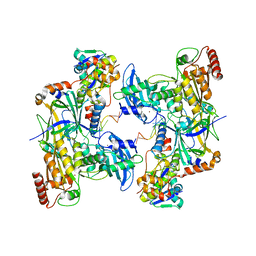 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 2 | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
8CX0
 
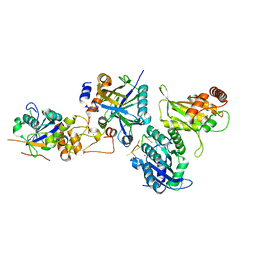 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC monomeric complex | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
8CX1
 
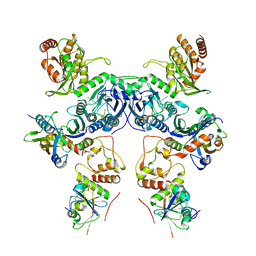 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 1 | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
7SEZ
 
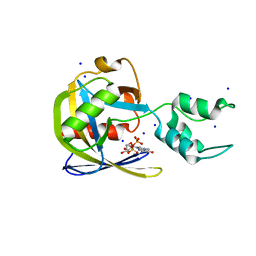 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, SODIUM ION | | Authors: | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.70001245 Å) | | Cite: | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
7SF0
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with trinucleotide substrate | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, MAGNESIUM ION, ... | | Authors: | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95000446 Å) | | Cite: | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
7T7H
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with inhibitor CP100356 | | Descriptor: | 4-(6,7-dimethoxy-3,4-dihydroisoquinolin-2(1H)-yl)-N-[2-(3,4-dimethoxyphenyl)ethyl]-6,7-dimethoxyquinazolin-2-amine, DNA repair NTP-phosphohydrolase, SODIUM ION | | Authors: | Peters, J.K, Gross, J.D. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78000259 Å) | | Cite: | Fluorescence-Based Activity Screening Assay Reveals Small Molecule Inhibitors of Vaccinia Virus mRNA Decapping Enzyme D9.
Acs Chem.Biol., 17, 2022
|
|
