3TEC
 
 | | CALCIUM BINDING TO THERMITASE. CRYSTALLOGRAPHIC STUDIES OF THERMITASE AT 0, 5 AND 100 MM CALCIUM | | Descriptor: | CALCIUM ION, EGLIN C, THERMITASE | | Authors: | Gros, P, Kalk, K.H, Hol, W.G.J. | | Deposit date: | 1990-10-26 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calcium binding to thermitase. Crystallographic studies of thermitase at 0, 5, and 100 mM calcium.
J.Biol.Chem., 266, 1991
|
|
2TEC
 
 | | MOLECULAR DYNAMICS REFINEMENT OF A THERMITASE-EGLIN-C COMPLEX AT 1.98 ANGSTROMS RESOLUTION AND COMPARISON OF TWO CRYSTAL FORMS THAT DIFFER IN CALCIUM CONTENT | | Descriptor: | CALCIUM ION, EGLIN C, THERMITASE | | Authors: | Gros, P, Betzel, C, Dauter, Z, Wilson, K.S, Hol, W.G.J. | | Deposit date: | 1990-10-26 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular dynamics refinement of a thermitase-eglin-c complex at 1.98 A resolution and comparison of two crystal forms that differ in calcium content.
J.Mol.Biol., 210, 1989
|
|
6H16
 
 | | Structure of LRP6 P3E3P4E4 in complex with VHH L-P2-D07 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Low-density lipoprotein receptor-related protein 6, ... | | Authors: | Gros, P, van Scherpenzeel, R.C. | | Deposit date: | 2018-07-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Anti-LRP5/6 VHHs promote differentiation of Wnt-hypersensitive intestinal stem cells.
Nat Commun, 10, 2019
|
|
6H15
 
 | | Structure of LRP6 P3E3P4E4 in complex with VHH L-P2-B10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Gros, P, van Scherpenzeel, R.C. | | Deposit date: | 2018-07-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Anti-LRP5/6 VHHs promote differentiation of Wnt-hypersensitive intestinal stem cells.
Nat Commun, 10, 2019
|
|
1TEC
 
 | | CRYSTALLOGRAPHIC REFINEMENT BY INCORPORATION OF MOLECULAR DYNAMICS. THE THERMOSTABLE SERINE PROTEASE THERMITASE COMPLEXED WITH EGLIN-C | | Descriptor: | CALCIUM ION, EGLIN C, SODIUM ION, ... | | Authors: | Gros, P, Dijkstra, B.W, Hol, W.G.J. | | Deposit date: | 1989-05-24 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic refinement by incorporation of molecular dynamics: thermostable serine protease thermitase complexed with eglin c.
Acta Crystallogr.,Sect.B, 45, 1989
|
|
4WX7
 
 | | Crystal structure of adenovirus 8 protease with a nitrile inhibitor | | Descriptor: | 3-[2-(3,5-dichlorophenyl)-2-methylpropanoyl]-N-(2-{[(2Z)-2-iminoethyl]amino}-2-oxoethyl)-4-methoxybenzamide, PVI, Protease | | Authors: | Grosche, P, Sirockin, F, Mac Sweeney, A, Ramage, P, Erbel, P, Melkko, S, Bernardi, A, Hughes, N, Ellis, D, Combrink, K, Jarousse, N, Altmann, E. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design and optimization of potent inhibitors of the adenoviral protease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4WX6
 
 | | Crystal structure of human adenovirus 8 protease with an irreversible vinyl sulfone inhibitor | | Descriptor: | N-[(2S)-2-(3,5-dichlorophenyl)-2-(ethylamino)acetyl]-3-methyl-L-valyl-N-[3-(methylsulfonyl)propyl]glycinamide, PVI, Protease | | Authors: | Grosche, P, Sirockin, F, Mac Sweeney, A, Ramage, P, Erbel, P, Melkko, S, Bernardi, A, Hughes, N, Ellis, D, Combrink, K, Jarousse, N, Altmann, E. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-based design and optimization of potent inhibitors of the adenoviral protease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4WX4
 
 | | Crystal structure of adenovirus 8 protease in complex with a nitrile inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCINE, N-[(2-cyanopyrimidin-4-yl)methyl]-3-[2-(3,5-dichlorophenyl)-2-methylpropanoyl]-4-methoxybenzamide, ... | | Authors: | Grosche, P, Sirockin, F, Mac Sweeney, A, Ramage, P, Erbel, P, Melkko, S, Bernardi, A, Hughes, N, Ellis, D, Combrink, K, Jarousse, N, Altmann, E. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure-based design and optimization of potent inhibitors of the adenoviral protease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1MPA
 
 | | BACTERICIDAL ANTIBODY AGAINST NEISSERIA MENINGITIDIS | | Descriptor: | CADMIUM ION, MN12H2 IGG2A-KAPPA, PORA P1.16 PEPTIDE FLUORESCEIN CONJUGATE | | Authors: | Van Den Elsen, J.M.H, Herron, J.N, Kroon, J, Gros, P. | | Deposit date: | 1997-02-26 | | Release date: | 1997-09-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bactericidal antibody recognition of a PorA epitope of Neisseria meningitidis: crystal structure of a Fab fragment in complex with a fluorescein-conjugated peptide.
Proteins, 29, 1997
|
|
8CML
 
 | |
6S08
 
 | | Crystal Structure of Properdin (TSR domains N1 & 456) | | Descriptor: | Properdin, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | van den Bos, R.M, Pearce, N.M, Gros, P. | | Deposit date: | 2019-06-14 | | Release date: | 2019-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Insights Into Enhanced Complement Activation by Structures of Properdin and Its Complex With the C-Terminal Domain of C3b.
Front Immunol, 10, 2019
|
|
6RLR
 
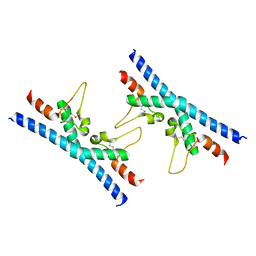 | | Crystal structure of CD9 large extracellular loop | | Descriptor: | CD9 antigen | | Authors: | Neviani, V, Kroon-Batenburg, L, Lutz, M, Pearce, N.M, Pos, W, Schotte, R, Spits, H, Gros, P. | | Deposit date: | 2019-05-02 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
3HRZ
 
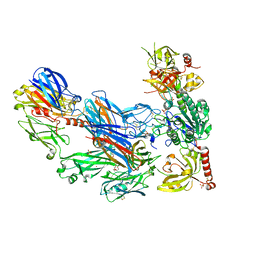 | | Cobra Venom Factor (CVF) in complex with human factor B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cobra venom factor, ... | | Authors: | Janssen, B.J.C, Gomes, L, Koning, R.I, Svergun, D.I, Koster, A.J, Fritzinger, D.C, Vogel, C.-W, Gros, P. | | Deposit date: | 2009-06-10 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into complement convertase formation based on the structure of the factor B-cobra venom factor complex
Embo J., 28, 2009
|
|
3HS0
 
 | | Cobra Venom Factor (CVF) in complex with human factor B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cobra venom factor, ... | | Authors: | Janssen, B.J.C, Gomes, L, Koning, R.I, Svergun, D.I, Koster, A.J, Fritzinger, D.C, Vogel, C.-W, Gros, P. | | Deposit date: | 2009-06-10 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into complement convertase formation based on the structure of the factor B-cobra venom factor complex
Embo J., 28, 2009
|
|
1FE8
 
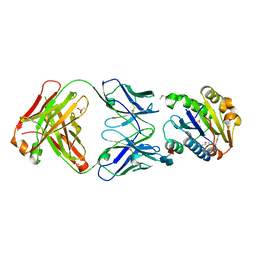 | | CRYSTAL STRUCTURE OF THE VON WILLEBRAND FACTOR A3 DOMAIN IN COMPLEX WITH A FAB FRAGMENT OF IGG RU5 THAT INHIBITS COLLAGEN BINDING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, IMMUNOGLOBULIN IGG RU5, ... | | Authors: | Bouma, B, Huizinga, E.G, Schiphorst, M.E, Sixma, J.J, Kroon, J, Gros, P. | | Deposit date: | 2000-07-21 | | Release date: | 2001-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of the collagen-binding site of the von Willebrand factor A3-domain.
J.Biol.Chem., 276, 2001
|
|
1ATZ
 
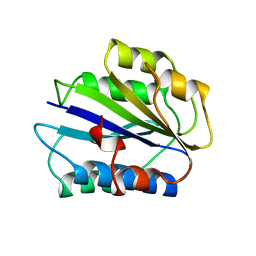 | | HUMAN VON WILLEBRAND FACTOR A3 DOMAIN | | Descriptor: | VON WILLEBRAND FACTOR | | Authors: | Huizinga, E.G, Gros, P. | | Deposit date: | 1997-08-15 | | Release date: | 1998-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the A3 domain of human von Willebrand factor: implications for collagen binding.
Structure, 5, 1997
|
|
3EF3
 
 | | cut-1a; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | (2,6-bis[(dimethylamino-kappaN)methyl]-4-{3-[(S)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}phenyl-kappaC~1~)(chloro)platinum(2+), Cutinase-1 | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-09-08 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
3ESD
 
 | | cut-2b; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | Cutinase 1, bromo(4-{3-[(R)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}-2,6-bis[(methylsulfanyl-kappaS)methyl]phenyl-kappaC~1~)palladium(2+) | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-10-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
3ESC
 
 | | cut-2a; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | Cutinase 1, bromo(4-{3-[(R)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}-2,6-bis[(methylsulfanyl-kappaS)methyl]phenyl-kappaC~1~)palladium(2+) | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-10-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
3ESA
 
 | | cut-1b; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | (2,6-bis[(dimethylamino-kappaN)methyl]-4-{3-[(S)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}phenyl-kappaC~1~)(chloro)platinum(2+), Cutinase 1 | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-10-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
3ESB
 
 | | cut-1c; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | (2,6-bis[(dimethylamino-kappaN)methyl]-4-{3-[(S)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}phenyl-kappaC~1~)(chloro)platinum(2+), CHLORIDE ION, Cutinase 1 | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-10-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
5FO8
 
 | | Crystal Structure of Human Complement C3b in Complex with MCP (CCP1-4) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3, ... | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
5FO7
 
 | | Crystal Structure of Human Complement C3b at 2.8 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3 BETA CHAIN, COMPLEMENT C3B ALPHA' CHAIN | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
5FO9
 
 | | Crystal Structure of Human Complement C3b in Complex with CR1 (CCP15- 17) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3 BETA CHAIN, COMPLEMENT C3B ALPHA' CHAIN, ... | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
5FOA
 
 | | Crystal Structure of Human Complement C3b in complex with DAF (CCP2-4) | | Descriptor: | COMPLEMENT C3 BETA CHAIN, COMPLEMENT C3B ALPHA CHAIN, DECAY ACCELERATING FACTOR, ... | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.188 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
