3KEF
 
 | | Crystal structure of IspH:DMAPP-complex | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, DIMETHYLALLYL DIPHOSPHATE, FE3-S4 CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KEM
 
 | | Crystal structure of IspH:IPP complex | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KE9
 
 | | Crystal structure of IspH:Intermediate-complex | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-24 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KEL
 
 | | Crystal Structure of IspH:PP complex | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, PYROPHOSPHATE 2- | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KE8
 
 | | Crystal structure of IspH:HMBPP-complex | | Descriptor: | 4-HYDROXY-3-METHYL BUTYL DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-24 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6EOZ
 
 | | Fe(II)/(alpha)ketoglutarate-dependent dioxygenase AsqJ_V72K mutant in complex with cyclopeptin (1b) | | Descriptor: | 2-OXOGLUTARIC ACID, Iron/alpha-ketoglutarate-dependent dioxygenase asqJ, NICKEL (II) ION, ... | | Authors: | Groll, M, Braeuer, A, Kaila, V.R.I. | | Deposit date: | 2017-10-10 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Catalytic mechanism and molecular engineering of quinolone biosynthesis in dioxygenase AsqJ.
Nat Commun, 9, 2018
|
|
1RYP
 
 | | CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | 20S PROTEASOME, MAGNESIUM ION | | Authors: | Groll, M, Ditzel, L, Loewe, J, Stock, D, Bochtler, M, Bartunik, H.D, Huber, R. | | Deposit date: | 1997-02-26 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of 20S proteasome from yeast at 2.4 A resolution.
Nature, 386, 1997
|
|
2R2Y
 
 | |
6Y36
 
 | | CCAAT-binding complex from Aspergillus fumigatus with cccA DNA | | Descriptor: | CCAAT-binding factor complex subunit HapC, CCAAT-binding factor complex subunit HapE, CCAAT-binding transcription factor subunit HAPB, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2020-02-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of HapE P88L -linked antifungal triazole resistance in Aspergillus fumigatus .
Life Sci Alliance, 3, 2020
|
|
6Y37
 
 | |
6Y35
 
 | | CCAAT-binding complex from Aspergillus fumigatus with cycA DNA | | Descriptor: | CCAAT-binding factor complex subunit HapC, CCAAT-binding factor complex subunit HapE, CCAAT-binding transcription factor subunit HAPB, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2020-02-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of HapE P88L -linked antifungal triazole resistance in Aspergillus fumigatus .
Life Sci Alliance, 3, 2020
|
|
6Y39
 
 | |
2GPL
 
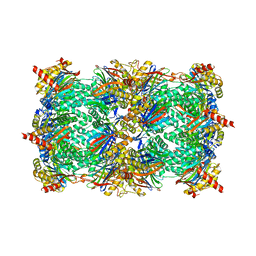 | | TMC-95 based biphenyl-ether macrocycles: specific proteasome inhibitors | | Descriptor: | BENZYL [12-(2-AMINO-2-OXOETHYL)-4-NITRO-10,13-DIOXO-15-[(PROPYLAMINO)CARBONYL]-2-OXA-11,14-DIAZATRICYCLO[15 .2.2.1~3,7~]DOCOSA-1(19),3(22),4,6,17,20-HEXAEN-9-YL]CARBAMATE, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Goetz, M, Kaiser, M, Weyher, E, Moroder, M. | | Deposit date: | 2006-04-18 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | TMC-95-Based Inhibitor Design Provides Evidence for the Catalytic Versatility of the Proteasome.
Chem.Biol., 13, 2006
|
|
3NZW
 
 | | Crystal structure of the yeast 20S proteasome in complex with 2b | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Gallastegui, N, Marechal, X, Le Ravalec, V, Basse, N, Richy, N, Genin, E, Huber, R, Moroder, M, Vidal, V, Reboud-Ravaux, M. | | Deposit date: | 2010-07-17 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 20S proteasome inhibition: designing noncovalent linear peptide mimics of the natural product TMC-95A.
Chemmedchem, 5, 2010
|
|
3NZJ
 
 | | Crystal structure of yeast 20S proteasome in complex with ligand 2a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Gallastegui, N, Marechal, X, Le Ravalec, V, Basse, N, Richy, N, Genin, E, Huber, R, Moroder, M, Vidal, V, Reboud-Ravaux, M. | | Deposit date: | 2010-07-16 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 20S proteasome inhibition: designing noncovalent linear peptide mimics of the natural product TMC-95A.
Chemmedchem, 5, 2010
|
|
3GPJ
 
 | | Crystal structure of the yeast 20S proteasome in complex with syringolin B | | Descriptor: | N-{[(1S)-2-methyl-1-{[(5S,8S)-5-(1-methylethyl)-2,7-dioxo-1,6-diazacyclododec-3-en-8-yl]carbamoyl}propyl]carbamoyl}-L-valine, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Huber, R, Kaiser, M. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic and structural studies on syringolin A and B reveal critical determinants of selectivity and potency of proteasome inhibition
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZCY
 
 | | yeast 20S proteasome:syringolin A-complex | | Descriptor: | (2S)-2-[[(2S)-1-[[(5S,8S,9E)-2,7-dioxo-5-propan-2-yl-1,6-diazacyclododeca-3,9-dien-8-yl]amino]-3-methyl-1-oxo-butan-2-yl]carbamoylamino]-3-methyl-butanoic acid, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Dudler, R, Kaiser, M. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A plant pathogen virulence factor inhibits the eukaryotic proteasome by a novel mechanism
Nature, 452, 2008
|
|
3HYE
 
 | | Crystal structure of 20S proteasome in complex with hydroxylated salinosporamide | | Descriptor: | (2R,3S,4R)-2-[(S)-(1S)-cyclohex-2-en-1-yl(hydroxy)methyl]-3-hydroxy-4-(2-hydroxyethyl)-3-methyl-5-oxopyrrolidine-2-carbaldehyde, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Arthur, K.A.M, Macherla, V.R, Manam, R.R, Potts, B.C. | | Deposit date: | 2009-06-22 | | Release date: | 2009-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Snapshots of the fluorosalinosporamide/20S complex offer mechanistic insights for fine tuning proteasome inhibition
J.Med.Chem., 52, 2009
|
|
3MHP
 
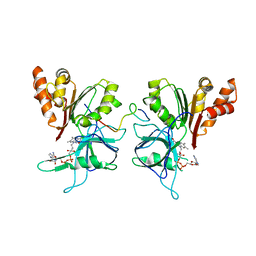 | | FNR-recruitment to the thylakoid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, leaf isozyme, ... | | Authors: | Groll, M, Alte, F, Soll, J, Boelter, B. | | Deposit date: | 2010-04-08 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ferredoxin:NADPH oxidoreductase is recruited to thylakoids by binding to a polyproline type II helix in a pH-dependent manner.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3D29
 
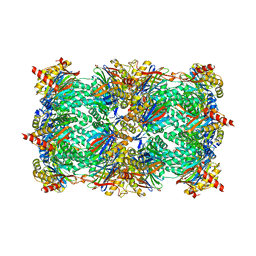 | | Proteasome Inhibition by Fellutamide B | | Descriptor: | (3R)-3-HYDROXYDODECANOIC ACID, Fellutamide B, PRE10 isoform 1, ... | | Authors: | Groll, M, Hines, J, Fahnestock, M, Crews, M.C. | | Deposit date: | 2008-05-07 | | Release date: | 2008-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Proteasome Inhibition by Fellutamide B Induces Nerve Growth Factor Synthesis
Chem.Biol., 15, 2008
|
|
3GPT
 
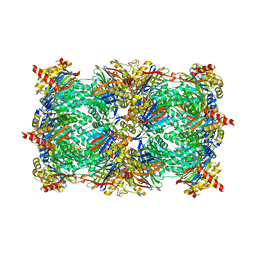 | | Crystal structure of the yeast 20S proteasome in complex with Salinosporamide derivatives: slow substrate ligand | | Descriptor: | (2R,3S,4R)-2-[(S)-(1S)-cyclohex-2-en-1-yl(hydroxy)methyl]-4-(2-fluoroethyl)-3-hydroxy-3-methyl-5-oxopyrrolidine-2-carbaldehyde, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Macherla, V.R, Manam, R.R, Arthur, K.A.M, Potts, C.B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Snapshots of the fluorosalinosporamide/20S complex offer mechanistic insights for fine tuning proteasome inhibition
J.Med.Chem., 52, 2009
|
|
3GPW
 
 | | Crystal structure of the yeast 20S proteasome in complex with Salinosporamide derivatives: irreversible inhibitor ligand | | Descriptor: | (3AR,6R,6AS)-6-((S)-((S)-CYCLOHEX-2-ENYL)(HYDROXY)METHYL)-6A-METHYL-4-OXO-HEXAHYDRO-2H-FURO[3,2-C]PYRROLE-6-CARBALDEHYDE, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Macherla, V.R, Manam, R.R, Arthur, K.A.M, Potts, C.B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Snapshots of the fluorosalinosporamide/20S complex offer mechanistic insights for fine tuning proteasome inhibition
J.Med.Chem., 52, 2009
|
|
3BDM
 
 | | yeast 20S proteasome:glidobactin A-complex | | Descriptor: | (2E,4E)-N-[(2S,3R)-3-hydroxy-1-[[(3Z,5S,8S,10S)-10-hydroxy-5-methyl-2,7-dioxo-1,6-diazacyclododec-3-en-8-yl]amino]-1-ox obutan-2-yl]dodeca-2,4-dienamide, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Dudler, R, Kaiser, M. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A plant pathogen virulence factor inhibits the eukaryotic proteasome by a novel mechanism
Nature, 452, 2008
|
|
3OKJ
 
 | | Alpha-keto-aldehyde binding mechanism reveals a novel lead structure motif for proteasome inhibition | | Descriptor: | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S,3S)-3-hydroxy-1-(4-hydroxyphenyl)-4-oxobutan-2-yl]-L-leucinamide, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Poynor, M, Gallastegui, P, Stein, M, Schmidt, B, Kloetzel, P.M, Huber, R. | | Deposit date: | 2010-08-25 | | Release date: | 2011-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Elucidation of the alpha-keto-aldehyde binding mechanism: a lead structure motif for proteasome inhibition
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3DY3
 
 | | Crystal structure of yeast 20S proteasome in complex with the epimer form of spirolactacystin | | Descriptor: | (3R,4R)-3-hydroxy-2-[(1S)-1-hydroxy-2-methylpropyl]-4-methyl-5-oxo-D-proline, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Balskus, E, Jacobsen, E. | | Deposit date: | 2008-07-25 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural analysis of spiro beta-lactone proteasome inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
