7NIL
 
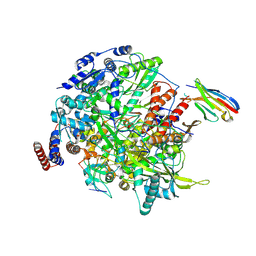 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8190 core | | Descriptor: | Nanobody8190 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.01 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
2J7N
 
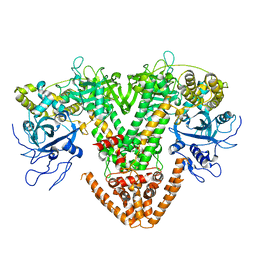 | | Structure of the RNAi polymerase from Neurospora crassa | | Descriptor: | GLYCEROL, MAGNESIUM ION, RNA-DEPENDENT RNA POLYMERASE | | Authors: | Salgado, P.S, Koivunen, M.R.L, Makeyev, E.V, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2006-10-13 | | Release date: | 2006-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of an Rnai Polymerase Links RNA Silencing and Transcription.
Plos Biol., 4, 2006
|
|
2JHP
 
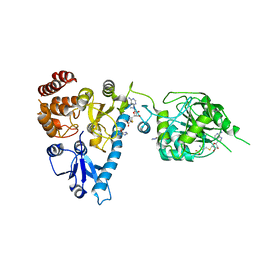 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | GUANINE, S-ADENOSYL-L-HOMOCYSTEINE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-23 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2JH9
 
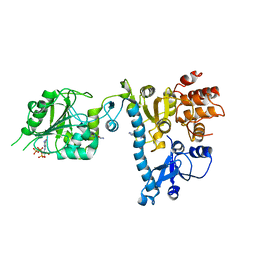 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | GUANINE, GUANOSINE-5'-TRIPHOSPHATE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2JH8
 
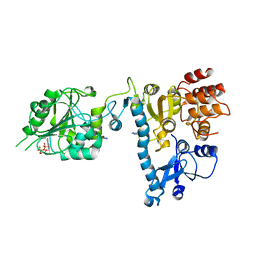 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, GUANINE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2W0C
 
 | | X-ray structure of the entire lipid-containing bacteriophage PM2 | | Descriptor: | CALCIUM ION, MAJOR CAPSID PROTEIN P2, PROTEIN 2, ... | | Authors: | Abrescia, N.G.A, Grimes, J.M, Kivela, H.M, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2008-08-13 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2
Mol.Cell, 31, 2008
|
|
2J7O
 
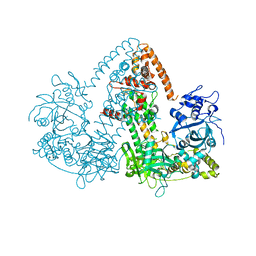 | | STRUCTURE OF THE RNAI POLYMERASE FROM NEUROSPORA CRASSA | | Descriptor: | MAGNESIUM ION, RNA DEPENDENT RNA POLYMERASE | | Authors: | Salgado, P.S, Koivunen, M.R.L, Makeyev, E.V, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2006-10-13 | | Release date: | 2006-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Structure of an Rnai Polymerase Links RNA Silencing and Transcription.
Plos Biol., 4, 2006
|
|
2JLG
 
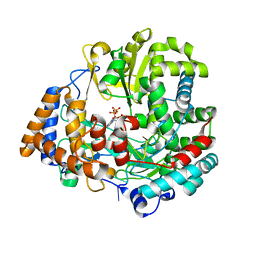 | | STRUCTURAL EXPLANATION FOR THE ROLE OF MN IN THE ACTIVITY OF PHI6 RNA-DEPENDENT RNA POLYMERASE | | Descriptor: | 5'-D(*DT DT DT DC DCP)-3', GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Poranen, M.M, Salgado, P.S, Koivunen, M.R.L, Wright, S, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2008-09-09 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Explanation for the Role of Mn2+ in the Activity of {Phi}6 RNA-Dependent RNA Polymerase.
Nucleic Acids Res., 36, 2008
|
|
2JLF
 
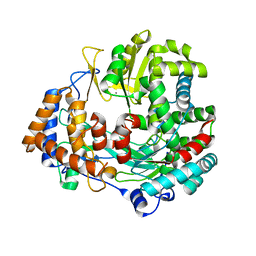 | | STRUCTURAL EXPLANATION FOR THE ROLE OF MN IN THE ACTIVITY OF PHI6 RNA- DEPENDENT RNA POLYMERASE | | Descriptor: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Poranen, M.M, Salgado, P.S, Koivunen, M.R.L, Wright, S, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2008-09-08 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Explanation for the Role of Mn2+ in the Activity of {Phi}6 RNA-Dependent RNA Polymerase.
Nucleic Acids Res., 36, 2008
|
|
2JHC
 
 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | GUANINE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2JL9
 
 | | Structural explanation for the role of Mn in the activity of phi6 RNA- dependent RNA polymerase | | Descriptor: | RNA-DIRECTED RNA POLYMERASE | | Authors: | Poranen, M.M, Salgado, P.S, Koivunen, M.R.L, Wright, S, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2008-09-05 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Explanation for the Role of Mn2+ in the Activity of {Phi}6 RNA-Dependent RNA Polymerase.
Nucleic Acids Res., 36, 2008
|
|
2JHA
 
 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | DIGUANOSINE-5'-TRIPHOSPHATE, GUANINE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2X7L
 
 | | Implications of the HIV-1 Rev dimer structure at 3.2A resolution for multimeric binding to the Rev response element | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, PROTEIN REV | | Authors: | DiMattia, M.A, Watts, N.R, Stahl, S.J, Rader, C, Wingfield, P.T, Stuart, D.I, Steven, A.C, Grimes, J.M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Implications of the HIV-1 Rev Dimer Structure at 3. 2 A Resolution for Multimeric Binding to the Rev Response Element.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2W2S
 
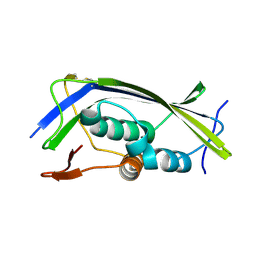 | | Structure of the Lagos bat virus matrix protein | | Descriptor: | MATRIX PROTEIN | | Authors: | Graham, S.C, Assenberg, R, Delmas, O, Verma, A, Gholami, A, Talbi, C, Owens, R.J, Stuart, D.I, Grimes, J.M, Bourhy, H. | | Deposit date: | 2008-11-03 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rhabdovirus Matrix Protein Structures Reveal a Novel Mode of Self-Association.
Plos Pathog., 4, 2008
|
|
2VGA
 
 | | The structure of Vaccinia virus A41 | | Descriptor: | PROTEIN A41 | | Authors: | Bahar, M.W, Kenyon, J.C, Putz, M.M, Abrescia, N.G.A, Pease, J.E, Wise, E.L, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Function of A41, a Vaccinia Virus Chemokine Binding Protein.
Plos Pathog., 4, 2008
|
|
2V8O
 
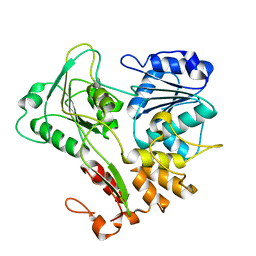 | | Structure of the Murray Valley encephalitis virus RNA helicase to 1. 9A resolution | | Descriptor: | FLAVIVIRIN PROTEASE NS3 | | Authors: | Mancini, E.J, Assenberg, R, Verma, A, Walter, T.S, Tuma, R, Grimes, J.M, Owens, R.J, Stuart, D.I. | | Deposit date: | 2007-08-09 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Murray Valley Encephalitis Virus RNA Helicase at 1.9 A Resolution.
Protein Sci., 16, 2007
|
|
2VWD
 
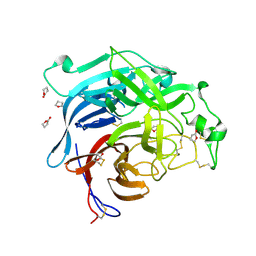 | | Nipah Virus Attachment Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GAMMA-BUTYROLACTONE, ... | | Authors: | Bowden, T.A, Crispin, M, Harvey, D.J, Aricescu, A.R, Grimes, J.M, Jones, E.Y, Stuart, D.I. | | Deposit date: | 2008-06-20 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure and Carbohydrate Analysis of Nipah Virus Attachment Glycoprotein: A Template for Antiviral and Vaccine Design.
J.Virol., 82, 2008
|
|
2VVY
 
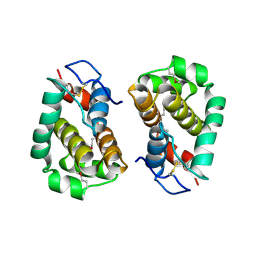 | | Structure of Vaccinia virus protein B14 | | Descriptor: | PROTEIN B15 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
2VVE
 
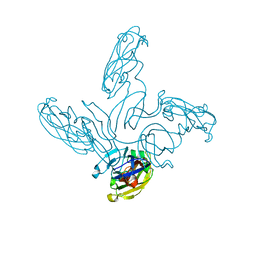 | | Crystal structure of the stem and receptor binding domain of the spike protein P1 from bacteriophage PM2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, SPIKE PROTEIN P1 | | Authors: | Abrescia, N.G.A, Grimes, J.M, Kivela, H.K, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2008-06-06 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2.
Mol.Cell, 31, 2008
|
|
2UXE
 
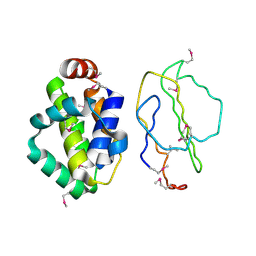 | | The structure of Vaccinia virus N1 | | Descriptor: | HYPOTHETICAL PROTEIN | | Authors: | Cooray, S, Bahar, M.W, Abrescia, N.G.A, McVey, C.E, Bartlett, N.W, Chen, R.A.-J, Stuart, D.I, Grimes, J.M, Smith, G.L. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functional and Structural Studies of the Vaccinia Virus Virulence Factor N1 Reveal a Bcl-2-Like Anti- Apoptotic Protein
J.Gen.Virol., 88, 2007
|
|
2UWI
 
 | | Structure of CrmE, a poxvirus TNF receptor | | Descriptor: | CRME PROTEIN | | Authors: | Graham, S.C, Bahar, M.W, Abrescia, N.G, Smith, G.L, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2007-03-22 | | Release date: | 2007-07-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Crme, a Virus-Encoded Tumour Necrosis Factor Receptor.
J.Mol.Biol., 372, 2007
|
|
2VHC
 
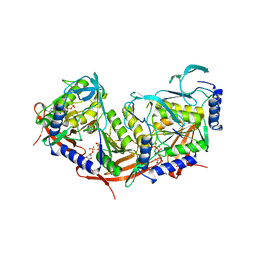 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 N234G mutant in complex with AMPCPP and MN | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
2VHQ
 
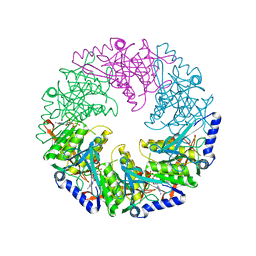 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 S252A mutant in complex with ATP AND MG | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-22 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
2VHJ
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 S252A mutant in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-21 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
2VVW
 
 | | Structure of Vaccinia virus protein A52 | | Descriptor: | PROTEIN A52 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
