4WR7
 
 | | Crystal structure of human carbonic anhydrase isozyme I with 2,3,5,6-Tetrafluoro-4-(propylthio)benzenesulfonamide. | | Descriptor: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-(propylsulfanyl)benzenesulfonamide, ACETATE ION, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2014-10-23 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Intrinsic thermodynamics of 4-substituted-2,3,5,6-tetrafluorobenzenesulfonamide binding to carbonic anhydrases by isothermal titration calorimetry.
Biophys.Chem., 205, 2015
|
|
6G5U
 
 | | Crystal structure of human carbonic anhydrase isozyme XIII with N-butyl-2,4-dichloro-5-sulfamoyl-benzamide | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Carbonic anhydrase 13, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2018-03-30 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of two-tail compounds with rotationally fixed benzenesulfonamide ring as inhibitors of carbonic anhydrases.
Eur J Med Chem, 156, 2018
|
|
6G6T
 
 | | Crystal structure of human carbonic anhydrase isozyme II with N-butyl-2,4-dichloro-5-sulfamoyl-benzamide | | Descriptor: | BICINE, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2018-04-03 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Design of two-tail compounds with rotationally fixed benzenesulfonamide ring as inhibitors of carbonic anhydrases.
Eur J Med Chem, 156, 2018
|
|
6G5L
 
 | | Crystal structure of human carbonic anhydrase isozyme XII with 4-chloro-2-(cyclohexylamino)-N-(2-hydroxyethyl)-5-sulfamoyl-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-(cyclohexylamino)-~{N}-(2-hydroxyethyl)-5-sulfamoyl-benzamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2018-03-29 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Design of two-tail compounds with rotationally fixed benzenesulfonamide ring as inhibitors of carbonic anhydrases.
Eur J Med Chem, 156, 2018
|
|
4WUP
 
 | | Crystal structure of human carbonic anhydrase isozyme I with 4-[(2-Hydroxyethyl)thio]benzenesulfonamide | | Descriptor: | 4-[(2-hydroxyethyl)sulfanyl]benzenesulfonamide, ACETATE ION, Carbonic anhydrase 1, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-11-03 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Intrinsic thermodynamics of 4-substituted-2,3,5,6-tetrafluorobenzenesulfonamide binding to carbonic anhydrases by isothermal titration calorimetry.
Biophys.Chem., 205, 2015
|
|
4WW8
 
 | | Crystal structure of human carbonic anhydrase isozyme XII with 4-Propylthiobenzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 4-(propylsulfanyl)benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-11-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Intrinsic thermodynamics of 4-substituted-2,3,5,6-tetrafluorobenzenesulfonamide binding to carbonic anhydrases by isothermal titration calorimetry.
Biophys.Chem., 205, 2015
|
|
6G7A
 
 | | Crystal structure of human carbonic anhydrase isozyme XII 2-(benzylamino)-4-chloro-N-(2-hydroxyethyl)-5-sulfamoyl-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-(2-hydroxyethyl)-2-[(phenylmethyl)amino]-5-sulfamoyl-benzamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2018-04-05 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Design of two-tail compounds with rotationally fixed benzenesulfonamide ring as inhibitors of carbonic anhydrases.
Eur J Med Chem, 156, 2018
|
|
2UYH
 
 | |
2GB7
 
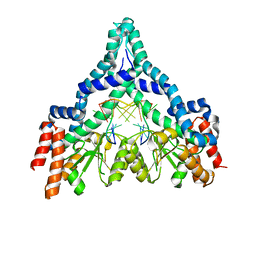 | | Metal-depleted Ecl18kI in complex with uncleaved, modified DNA | | Descriptor: | DNA STRAND 1, DNA STRAND 2, R.Ecl18kI | | Authors: | Bochtler, M, Szczepanowski, R.H, Tamulaitis, G, Grazulis, S, Czapinska, H, Manakova, E, Siksnys, V. | | Deposit date: | 2006-03-10 | | Release date: | 2006-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease
Embo J., 25, 2006
|
|
2FQZ
 
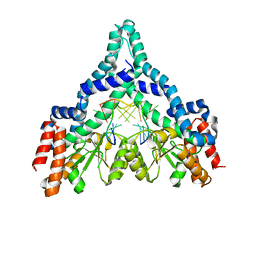 | | Metal-depleted Ecl18kI in complex with uncleaved DNA | | Descriptor: | DNA STRAND 1, DNA STRAND 2, R.Ecl18kI | | Authors: | Bochtler, M, Szczepanowski, R.H, Tamulaitis, G, Grazulis, S, Czapinska, H, Manakova, E, Siksnys, V. | | Deposit date: | 2006-01-18 | | Release date: | 2006-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease
Embo J., 25, 2006
|
|
2VLA
 
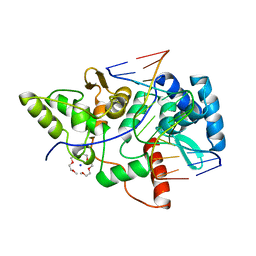 | | Crystal structure of restriction endonuclease BpuJI recognition domain in complex with cognate DNA | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 5'-D(*GP*GP*TP*AP*CP*CP*CP*GP*TP*GP *GP*A)-3', 5'-D(*TP*CP*CP*AP*CP*GP*GP*GP*TP*AP *CP*C)-3', ... | | Authors: | Sukackaite, R, Grazulis, S, Bochtler, M, Siksnys, V. | | Deposit date: | 2008-01-11 | | Release date: | 2008-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Recognition Domain of the Bpuji Restriction Endonuclease in Complex with Cognate DNA at 1.3-A Resolution.
J.Mol.Biol., 378, 2008
|
|
2C7Q
 
 | |
2C7O
 
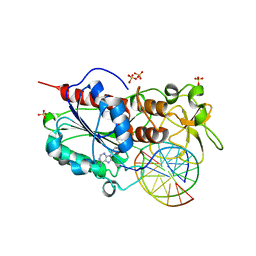 | |
2UYC
 
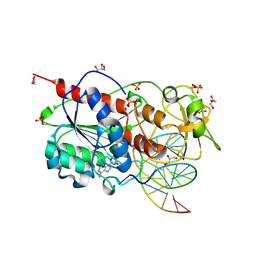 | |
2UZ4
 
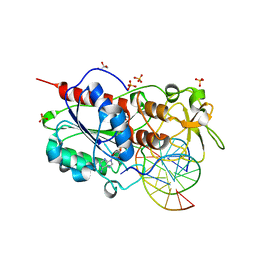 | |
2C7P
 
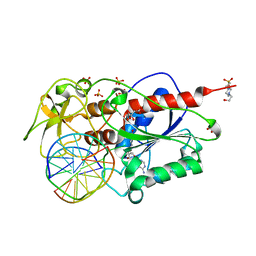 | | HhaI DNA methyltransferase complex with oligonucleotide containing 2- aminopurine opposite to the target base (GCGC:GMPC) and SAH | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-D(*G*GP*AP*TP*GP*(5CM*2PR)*CP*TP*GP*AP*C)-3', 5'-D(*G*TP*CP*AP*GP*CP*GP*CP*AP*TP*CP*C)-3', ... | | Authors: | Neely, R.K, Daujotyte, D, Grazulis, S, Magennis, S.W, Dryden, D.T.F, Klimasauskas, S, Jones, A.C. | | Deposit date: | 2005-11-25 | | Release date: | 2005-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Time-Resolved Fluorescence of 2-Aminopurine as a Probe of Base Flipping in M.HhaI-DNA Complexes.
Nucleic Acids Res., 33, 2005
|
|
2C7R
 
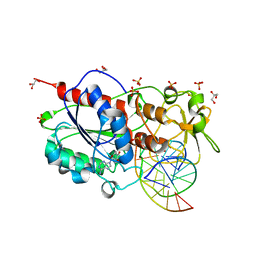 | |
2IXS
 
 | | Structure of SdaI restriction endonuclease | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SDAI RESTRICTION ENDONUCLEASE, ... | | Authors: | Tamulaitiene, G, Jakubauskas, A, Urbanke, C, Huber, R, Grazulis, S, Siksnys, V. | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Rare-Cutting Restriction Enzyme Sdai Reveals Unexpected Domain Architecture
Structure, 14, 2006
|
|
5MSB
 
 | | Crystal structure of human carbonic anhydrase isozyme XII with 2,3,5,6-tetrafluoro-4[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2017-01-02 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure correlations with the intrinsic thermodynamics of human carbonic anhydrase inhibitor binding.
PeerJ, 6, 2018
|
|
5OGO
 
 | | Crystal structure of chimeric carbonic anhydrase I with 3-(Benzylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 3-(benzylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, BICINE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2017-07-13 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Crystal structure of chimeric carbonic anhydrase I with 3-(Benzylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide
To be published
|
|
5OGJ
 
 | | Crystal structure of human carbonic anhydrase isozyme XIII with 2-(Cyclooctylamino)-3,5,6-trifluoro-4-[(2-hydroxyethyl)thio]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2-(cyclooctylamino)-3,5,6-trifluoro-4-[(2-hydroxyethyl)sulfanyl]benzenesulfonamide, ACETATE ION, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2017-07-13 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Crystal structure of human carbonic anhydrase isozyme XIII with 2-(Cyclooctylamino)-3,5,6-trifluoro-4-[(2-hydroxyethyl)thio]benzenesulfonamide
To be published
|
|
5OHH
 
 | | Crystal structure of human carbonic anhydrase isozyme XIII with 2-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)thio]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[(1S)-2,3-dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)sulfanyl]benzenesulfonamide, ACETATE ION, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2017-07-17 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of human carbonic anhydrase isozyme XIII with 2-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)thio]benzenesulfonamide
To be published
|
|
5MSA
 
 | | Crystal structure of human carbonic anhydrase isozyme XII with 2,3,5,6-Tetrafluoro-4-(propylthio)benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-(propylsulfanyl)benzenesulfonamide, CITRIC ACID, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2017-01-02 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of human carbonic anhydrase
isozyme XII with 2,3,5,6-Tetrafluoro-4-(propylthio)benzenesulfonamide
To be published
|
|
7PUW
 
 | | Crystal structure of carbonic anhydrase XII with methyl 2-chloro-4-[(2-phenylethyl)sulfanyl]-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 12, ZINC ION, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
7PUU
 
 | | Crystal structure of carbonic anhydrase XII with methyl 4-chloro-2-cyclohexylsulfanyl-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 12, ZINC ION, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
