1VBA
 
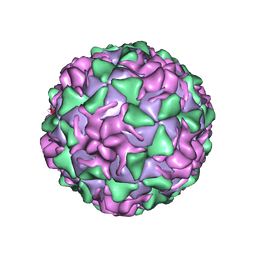 | | POLIOVIRUS (TYPE 3, SABIN STRAIN) (P3/SABIN, P3/LEON/12A(1)B) COMPLEXED WITH R78206 | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE PROPYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 3 | | Authors: | Grant, R.A, Hiremath, C.N, Filman, D.J, Syed, R, Andries, K, Hogle, J.M. | | Deposit date: | 1996-01-02 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design.
Curr.Biol., 4, 1994
|
|
1VBB
 
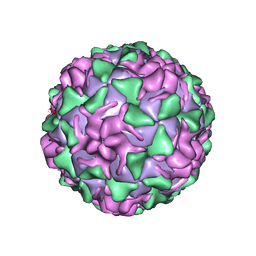 | | POLIOVIRUS (TYPE 3, SABIN STRAIN) (P3/SABIN, P3/LEON/12A(1)B) COMPLEXED WITH R80633 | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE BUTYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 3 | | Authors: | Grant, R.A, Hiremath, C.N, Filman, D.J, Syed, R, Andries, K, Hogle, J.M. | | Deposit date: | 1996-01-02 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design.
Curr.Biol., 4, 1994
|
|
1VBC
 
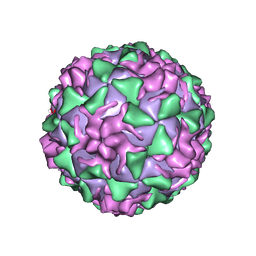 | | POLIOVIRUS (TYPE 3, SABIN STRAIN) (P3/SABIN, P3/LEON/12A(1)B) COMPLEXED WITH R77975 | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE ETHYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 3 | | Authors: | Grant, R.A, Hiremath, C.N, Filman, D.J, Syed, R, Andries, K, Hogle, J.M. | | Deposit date: | 1996-01-02 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design.
Curr.Biol., 4, 1994
|
|
1VBD
 
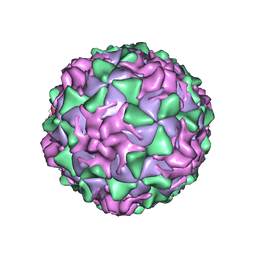 | | POLIOVIRUS (TYPE 1, MAHONEY STRAIN) COMPLEXED WITH R78206 | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE PROPYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 1 MAHONEY | | Authors: | Grant, R.A, Hiremath, C.N, Filman, D.J, Syed, R, Andries, K, Hogle, J.M. | | Deposit date: | 1996-01-02 | | Release date: | 1996-07-11 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design.
Curr.Biol., 4, 1994
|
|
1VBE
 
 | | POLIOVIRUS (TYPE 3, SABIN STRAIN, MUTANT 242-H2) COMPLEXED WITH R78206 | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE PROPYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 3 | | Authors: | Grant, R.A, Hiremath, C.N, Filman, D.J, Syed, R, Andries, K, Hogle, J.M. | | Deposit date: | 1996-01-02 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design.
Curr.Biol., 4, 1994
|
|
3B8J
 
 | |
1DU0
 
 | | ENGRAILED HOMEODOMAIN Q50A VARIANT DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*GP*GP*TP*AP*AP*TP*TP*AP*CP*AP*TP*GP*GP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*TP*AP*CP*CP*TP*AP*A)-3'), ENGRAILED HOMEODOMAIN | | Authors: | Grant, R.A, Rould, M.A, Klemm, J.D, Pabo, C.O. | | Deposit date: | 2000-01-13 | | Release date: | 2000-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the role of glutamine 50 in the homeodomain-DNA interface: crystal structure of engrailed (Gln50 --> ala) complex at 2.0 A.
Biochemistry, 39, 2000
|
|
1DPS
 
 | | THE CRYSTAL STRUCTURE OF DPS, A FERRITIN HOMOLOG THAT BINDS AND PROTECTS DNA | | Descriptor: | DPS, SODIUM ION | | Authors: | Grant, R.A, Filman, D.J, Finkel, S.E, Kolter, R, Hogle, J.M. | | Deposit date: | 1998-02-23 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Dps, a ferritin homolog that binds and protects DNA.
Nat.Struct.Biol., 5, 1998
|
|
1TME
 
 | | THREE-DIMENSIONAL STRUCTURE OF THEILER VIRUS | | Descriptor: | THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP1), THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP2), THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP3), ... | | Authors: | Grant, R.A, Filman, D.J, Hogle, J.M. | | Deposit date: | 1992-01-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of Theiler virus.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
2PIE
 
 | |
5JI2
 
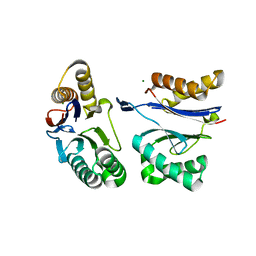 | | HslU L199Q in HslUV complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV, ... | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.307 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
5JI3
 
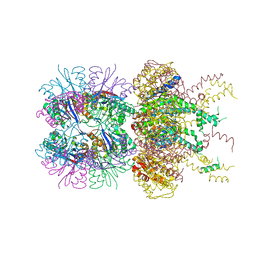 | | HslUV complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
6OTT
 
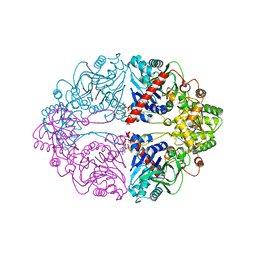 | | Structure of PurF in complex with ppApp | | Descriptor: | Amidophosphoribosyltransferase, PurF, MAGNESIUM ION, ... | | Authors: | Grant, R.A, Wang, B, Laub, M.T. | | Deposit date: | 2019-05-03 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp.
Nature, 575, 2019
|
|
4X9V
 
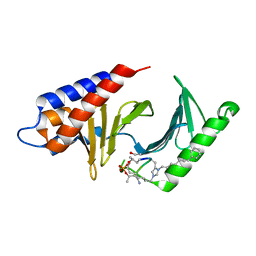 | | PLK-1 polo-box domain in complex with Bioactive Imidazolium-containing phosphopeptide macrocycle 3C | | Descriptor: | Phosphopeptide macrocycle 3C, Serine/threonine-protein kinase PLK1 | | Authors: | Grant, R.A, Qian, W.-J, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2014-12-11 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | Neighbor-directed histidine N ( tau )-alkylation: A route to imidazolium-containing phosphopeptide macrocycles.
Biopolymers, 104, 2015
|
|
4X9W
 
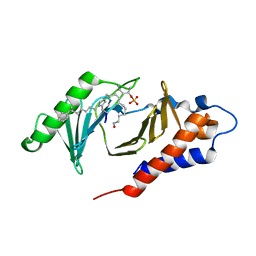 | | PLK-1 polo-box domain in complex with Bioactive Imidazolium-containing phosphopeptide macrocycle 4C | | Descriptor: | Macrocyclic phosphopeptide 4C, Serine/threonine-protein kinase PLK1 | | Authors: | Grant, R.A, Qian, W.-J, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2014-12-11 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Neighbor-directed histidine N ( tau )-alkylation: A route to imidazolium-containing phosphopeptide macrocycles.
Biopolymers, 104, 2015
|
|
4X9R
 
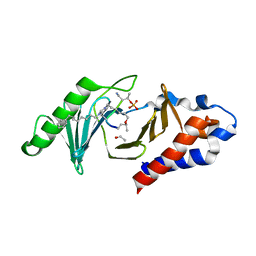 | | PLK-1 polo-box domain in complex with Bioactive Imidazolium-containing phosphopeptide macrocycle 3B | | Descriptor: | Phosphopeptide macrocycle 3B, Serine/threonine-protein kinase PLK1 | | Authors: | Grant, R.A, Qian, W.-J, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2014-12-11 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Neighbor-directed histidine N ( tau )-alkylation: A route to imidazolium-containing phosphopeptide macrocycles.
Biopolymers, 104, 2015
|
|
7MX1
 
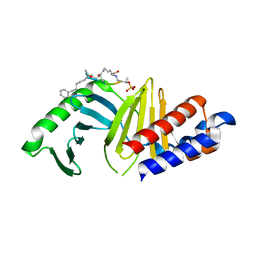 | | PLK-1 polo-box domain in complex with a high affinity macrocycle synthesized using a novel glutamic acid analog | | Descriptor: | ACE-PRO-LEU-ALA-SER-TPO, N-[(4S)-4,5-diamino-5-oxopentyl]-10-phenyldecanamide, Serine/threonine-protein kinase PLK1 | | Authors: | Grant, R.A, Hymel, D, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2021-05-17 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design and synthesis of a new orthogonally protected glutamic acid analog and its use in the preparation of high affinity polo-like kinase 1 polo-box domain - binding peptide macrocycles.
Org.Biomol.Chem., 19, 2021
|
|
6AX4
 
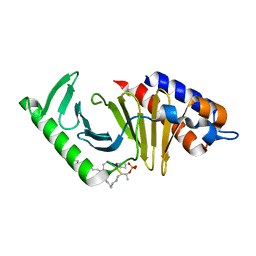 | | Plk-1 polo-box domain in complex with histidine N(tau)-cyclized Macrocycle 5b. | | Descriptor: | AMYLAMINE, Serine/threonine-protein kinase PLK1, histidine N(tau)-cyclized Macrocycle 5b | | Authors: | Grant, R.A, Hymel, D, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2017-09-06 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Histidine N( tau )-cyclized macrocycles as a new genre of polo-like kinase 1 polo-box domain-binding inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
5TXV
 
 | | HslU P21 cell with 4 hexamers | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU | | Authors: | Grant, R.A, Chen, J, Glynn, S.E, Sauer, R.T. | | Deposit date: | 2016-11-17 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (7.086 Å) | | Cite: | Covalently linked HslU hexamers support a probabilistic mechanism that links ATP hydrolysis to protein unfolding and translocation.
J. Biol. Chem., 292, 2017
|
|
1PO2
 
 | | POLIOVIRUS (TYPE 1, MAHONEY) IN COMPLEX WITH R77975, AN INHIBITOR OF VIRAL REPLICATION | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE ETHYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 1 MAHONEY | | Authors: | Hiremath, C.N, Filman, D.J, Grant, R.A, Hogle, J.M. | | Deposit date: | 1997-01-08 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-induced conformational changes in poliovirus-antiviral drug complexes.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1PO1
 
 | | POLIOVIRUS (TYPE 1, MAHONEY) IN COMPLEX WITH R80633, AN INHIBITOR OF VIRAL REPLICATION | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE BUTYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 1 MAHONEY | | Authors: | Hiremath, C.N, Filman, D.J, Grant, R.A, Hogle, J.M. | | Deposit date: | 1997-01-08 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-induced conformational changes in poliovirus-antiviral drug complexes.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1PIV
 
 | | BINDING OF THE ANTIVIRAL DRUG WIN51711 TO THE SABIN STRAIN OF TYPE 3 POLIOVIRUS: STRUCTURAL COMPARISON WITH DRUG BINDING IN RHINOVIRUS 14 | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, POLIOVIRUS TYPE 3 (SUBUNIT VP1), ... | | Authors: | Hiremath, C.N, Grant, R.A, Filman, D.J, Hogle, J.M. | | Deposit date: | 1995-02-02 | | Release date: | 1995-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Binding of the antiviral drug WIN51711 to the sabin strain of type 3 poliovirus: structural comparison with drug binding in rhinovirus 14.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1TWB
 
 | | SspB disulfide crosslinked to an ssrA degradation tag | | Descriptor: | Stringent starvation protein B homolog, ssrA peptide | | Authors: | Bolon, D.N, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide-Dependent Substrate Handoff from the SspB Adaptor to the AAA+ ClpXP Protease.
Mol.Cell, 16, 2004
|
|
1U9P
 
 | | Permuted single-chain Arc | | Descriptor: | pArc | | Authors: | Tabtiang, R.K, Cezairliyan, B.O, Grant, R.A, Cochrane, J.C, Sauer, R.T. | | Deposit date: | 2004-08-10 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Consolidating critical binding determinants by noncyclic rearrangement of protein secondary structure
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2PQK
 
 | | X-ray crystal structure of human Mcl-1 in complex with Bim BH3 | | Descriptor: | Bim BH3 peptide, Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Bare, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2007-05-02 | | Release date: | 2007-06-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
