1QVV
 
 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
1QVW
 
 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | GLYCEROL, YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
1HEZ
 
 | | Structure of P. magnus protein L bound to a human IgM Fab. | | Descriptor: | HEAVY CHAIN OF IG, IMIDAZOLE, KAPPA LIGHT CHAIN OF IG, ... | | Authors: | Graille, M, Stura, E.A, Housden, N.G, Bottomley, S.P, Taussig, M.J, Sutton, B.J, Gore, M.G, Charbonnier, J.B. | | Deposit date: | 2000-11-27 | | Release date: | 2001-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex between Peptostreptococcus Magnus Protein L and a Human Antibody Reveals Structural Convergence in the Interaction Modes of Fab Binding Modes
Structure, 9, 2001
|
|
1MHH
 
 | |
1DEE
 
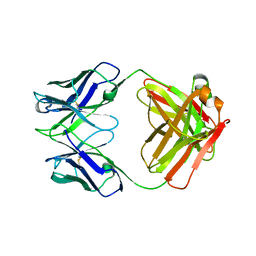 | | Structure of S. aureus protein A bound to a human IgM Fab | | Descriptor: | IGM RF 2A2, IMMUNOGLOBULIN G BINDING PROTEIN A | | Authors: | Graille, M, Stura, E.A, Corper, A.L, Sutton, B.J, Taussig, M.J, Charbonnier, J.B, Silverman, G.J. | | Deposit date: | 1999-11-15 | | Release date: | 2000-05-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2ZBK
 
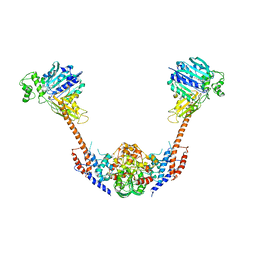 | | Crystal structure of an intact type II DNA topoisomerase: insights into DNA transfer mechanisms | | Descriptor: | RADICICOL, Type 2 DNA topoisomerase 6 subunit B, Type II DNA topoisomerase VI subunit A | | Authors: | Graille, M, Cladiere, L, Durand, D, Lecointe, F, Forterre, P, van Tilbeurgh, H, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2007-10-22 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Crystal Structure of an Intact Type II DNA Topoisomerase: Insights into DNA Transfer Mechanisms
Structure, 16, 2008
|
|
3M7P
 
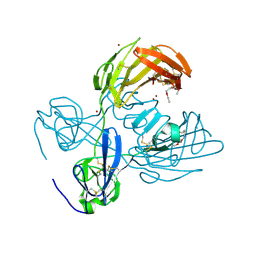 | | Fibronectin fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DODECAETHYLENE GLYCOL, FN1 protein, ... | | Authors: | Graille, M, Pagano, M, Rose, T, Reboud Ravaux, M, van Tilbeurgh, H. | | Deposit date: | 2010-03-17 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Zinc Induces Structural Reorganization of Gelatin Binding Domain from Human Fibronectin and Affects Collagen Binding
Structure, 18, 2010
|
|
6YYL
 
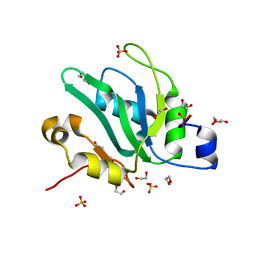 | | Crystal structure of S. pombe Mei2 RRM3 domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Meiosis protein mei2, ... | | Authors: | Graille, M, Hazra, D. | | Deposit date: | 2020-05-05 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A scaffold lncRNA shapes the mitosis to meiosis switch.
Nat Commun, 12, 2021
|
|
6Y3Z
 
 | |
6Y3P
 
 | |
1YNT
 
 | | Structure of the monomeric form of T. gondii SAG1 surface antigen bound to a human Fab | | Descriptor: | 4F11E12 Fab variable heavy chain region, 4F11E12 Fab variable light chain region, CADMIUM ION, ... | | Authors: | Graille, M, Stura, E.A, Bossus, M, Muller, B.H, Letourneur, O, Battail-Poirot, N, Sibai, G, Rolland, D, Le Du, M.H, Ducancel, F. | | Deposit date: | 2005-01-25 | | Release date: | 2005-12-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the complex between the monomeric form of Toxoplasma gondii surface antigen 1 (SAG1) and a monoclonal antibody that mimics the human immune response
J.Mol.Biol., 354, 2005
|
|
6F5Z
 
 | |
2VGM
 
 | |
2VGN
 
 | | Structure of S. cerevisiae Dom34, a translation termination-like factor involved in RNA quality control pathways and interacting with Hbs1 (SelenoMet-labeled protein) | | Descriptor: | DOM34, GLYCEROL, PHOSPHATE ION | | Authors: | Graille, M, Chaillet, M, Van Tilbeurgh, H. | | Deposit date: | 2007-11-14 | | Release date: | 2008-01-22 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Structure of Yeast Dom34: A Protein Related to Translation Termination Factor Erf1 and Involved in No-Go Decay.
J.Biol.Chem., 283, 2008
|
|
1QVZ
 
 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
1XTZ
 
 | | Crystal structure of the S. cerevisiae D-ribose-5-phosphate isomerase: comparison with the archeal and bacterial enzymes | | Descriptor: | Ribose-5-phosphate isomerase | | Authors: | Graille, M, Meyer, P, Leulliot, N, Sorel, I, Janin, J, van Tilbeurgh, H, Quevillon-Cheruel, S. | | Deposit date: | 2004-10-25 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the S. cerevisiae D-ribose-5-phosphate isomerase: comparison with the archaeal and bacterial enzymes
Biochimie, 87, 2005
|
|
6ZXW
 
 | |
6ZXV
 
 | |
6ZXY
 
 | |
1V74
 
 | | Structure of the E. coli colicin D bound to its immunity protein ImmD | | Descriptor: | Colicin D, Colicin D immunity protein, PENTAETHYLENE GLYCOL | | Authors: | Graille, M, Mora, L, Buckingham, R.H, van Tilbeurgh, H, de Zamaroczy, M. | | Deposit date: | 2003-12-10 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural inhibition of the colicin D tRNase by the tRNA-mimicking immunity protein
Embo J., 23, 2004
|
|
1TLV
 
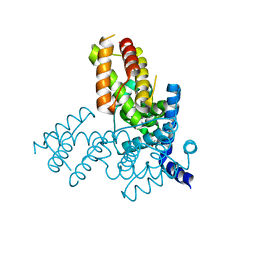 | | Structure of the native and inactive LicT PRD from B. subtilis | | Descriptor: | Transcription antiterminator licT | | Authors: | Graille, M, Zhou, C.-Z, Receveur-Brechot, V, Collinet, B, Declerck, N, van Tilbeurgh, H. | | Deposit date: | 2004-06-10 | | Release date: | 2005-02-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Activation of the LicT Transcriptional Antiterminator Involves a Domain Swing/Lock Mechanism Provoking Massive Structural Changes
J.Biol.Chem., 280, 2005
|
|
2CIS
 
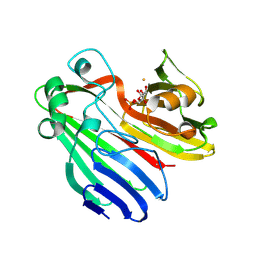 | | Structure-based functional annotation: Yeast ymr099c codes for a D- hexose-6-phosphate mutarotase. Complex with tagatose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-tagatofuranose, BARIUM ION, GLUCOSE-6-PHOSPHATE 1-EPIMERASE | | Authors: | Graille, M, Baltaze, J.-P, Leulliot, N, Liger, D, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2006-03-24 | | Release date: | 2006-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-based functional annotation: yeast ymr099c codes for a D-hexose-6-phosphate mutarotase.
J. Biol. Chem., 281, 2006
|
|
2CIR
 
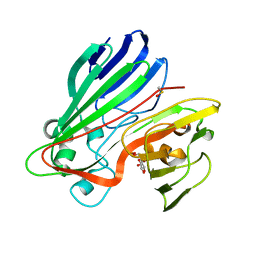 | | Structure-based functional annotation: Yeast ymr099c codes for a D- hexose-6-phosphate mutarotase. Complex with glucose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, HEXOSE-6-PHOSPHATE MUTAROTASE | | Authors: | Graille, M, Baltaze, J.-P, Leulliot, N, Liger, D, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2006-03-24 | | Release date: | 2006-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based functional annotation: yeast ymr099c codes for a D-hexose-6-phosphate mutarotase.
J. Biol. Chem., 281, 2006
|
|
2CIQ
 
 | | Structure-based functional annotation: Yeast ymr099c codes for a D- hexose-6-phosphate mutarotase. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, HEXOSE-6-PHOSPHATE MUTAROTASE, ... | | Authors: | Graille, M, Baltaze, J.-P, Leulliot, N, Liger, D, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2006-03-24 | | Release date: | 2006-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based functional annotation: yeast ymr099c codes for a D-hexose-6-phosphate mutarotase.
J. Biol. Chem., 281, 2006
|
|
2B3T
 
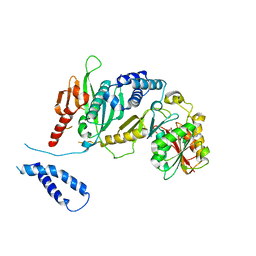 | | Structure of complex between E. coli translation termination factor RF1 and the PrmC methyltransferase | | Descriptor: | Peptide chain release factor 1, Protein methyltransferase hemK, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Graille, M, Heurgue-Hamard, V, Champ, S, Mora, L, Scrima, N, Ulryck, N, van Tilbeurgh, H, Buckingham, R.H. | | Deposit date: | 2005-09-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular basis for bacterial class I release factor methylation by PrmC
Mol.Cell, 20, 2005
|
|
