5MWL
 
 | | INOSITOL 1,3,4,5,6-PENTAKISPHOSPHATE 2-KINASE FROM M. MUSCULUS IN COMPLEX WITH ATP and IP5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inositol-pentakisphosphate 2-kinase, MAGNESIUM ION, ... | | Authors: | Franco-Echevarria, E, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of mammalian inositol 1,3,4,5,6-pentakisphosphate 2-kinase reveals a new zinc-binding site and key features for protein function.
J. Biol. Chem., 292, 2017
|
|
1E4I
 
 | | 2-deoxy-2-fluoro-beta-D-glucosyl/enzyme intermediate complex of the beta-glucosidase from Bacillus polymyxa | | Descriptor: | 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, 2-deoxy-2-fluoro-alpha-D-glucopyranose, BETA-GLUCOSIDASE | | Authors: | Sanz-Aparicio, J, Gonzalez, B, Hermoso, J.A, Arribas, J.C, Canada, F.J, Polaina, J. | | Deposit date: | 2000-07-06 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Increased Resistance to Thermal Denaturation Induced by Single Amino Acid Substitution in the Sequence of Beta-Glucosidase a from Bacillus Polymyxa.
Proteins: Struct.,Funct., Genet., 33, 1998
|
|
5O1Z
 
 | |
5O1W
 
 | |
5O20
 
 | | Structure of Nrd1 RNA binding domain in complex with RNA (UUAGUAAUCC) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein NRD1, RNA (5'-R(*UP*AP*GP*UP*AP*AP*UP*C)-3') | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
5O1Y
 
 | | Structure of Nrd1 RNA binding domain in complex with RNA (GUAA) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein NRD1, ... | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
5O1X
 
 | | Structure of Nrd1 RNA binding domain | | Descriptor: | 1,2-ETHANEDIOL, Protein NRD1, THIOCYANATE ION | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
8PPG
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with beta-D-glucopyranosyl 1,3,4,6-tetrakisphosphate/ADP/Mn after reaction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol-trisphosphate 3-kinase A, MANGANESE (II) ION, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPE
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with DL-6-deoxy-6-phosphoryloxymethyl-scyllo-inositol 1,2,4-trisphosphate/ADP/Mn | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DL-6-deoxy-6-phosphoryloxymethyl-scyllo-inositol 1,2,4-trisphosphate, GLYCEROL, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPB
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with D-3-deoxy-myo-inositol 1,4,6-trisphosphate/ATP/Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-3-deoxy-myo-inositol 1,4,6-trisphosphate, Inositol-trisphosphate 3-kinase A, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPC
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with L-chiro-inositol 2,3,5-trisphosphate/ATP/Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inositol-trisphosphate 3-kinase A, L-chiro-inositol 2,3,5-trisphosphate, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPJ
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with beta-D-glucopyranosylmethanol 3,4,6,1'-tetrakisphosphate/ADP/Mn after reaction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol-trisphosphate 3-kinase A, MANGANESE (II) ION, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPD
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with DL-6-deoxy-6-hydroxy-methyl-scyllo-inositol 1,2,4-trisphosphate/ATP/Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DL-6-deoxy-6-hydroxy-methyl-scyllo-inositol 1,2,4-trisphosphate, Inositol-trisphosphate 3-kinase A, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPH
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with alpha-D-glucopyranosyl 1,3,4-trisphosphate/ATP/Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inositol-trisphosphate 3-kinase A, MANGANESE (II) ION, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PP9
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with scyllo-inositol 1,2,3,5-tetrakisphosphate/ADP/Mn | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol-trisphosphate 3-kinase A, MANGANESE (II) ION, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPI
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with beta-D-glucopyranosylmethanol 3,4,1'-trisphosphate/ATP/Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inositol-trisphosphate 3-kinase A, MANGANESE (II) ION, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPA
 
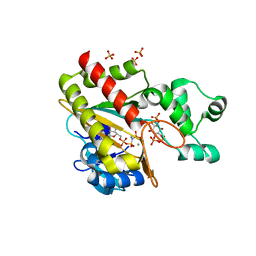 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with D-myo-inositol 1,4,6-trisphosphate/AMP-PNP/Mn | | Descriptor: | D-myo-inositol 1,4,6-trisphosphate, Inositol-trisphosphate 3-kinase A, MANGANESE (II) ION, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PP8
 
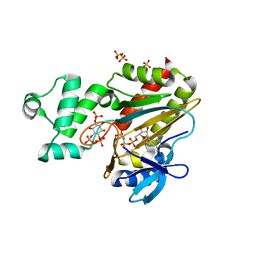 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with L-scyllo-inositol 1,2,4-trisphosphate/AMP-PNP/Mn | | Descriptor: | Inositol-trisphosphate 3-kinase A, L-scyllo-inositol 1,2,4-trisphosphate, MANGANESE (II) ION, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPF
 
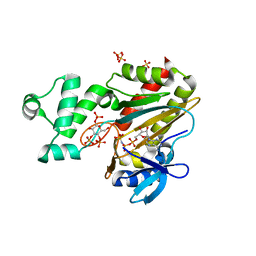 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with beta-D-glucopyranosyl 1,3,4-trisphosphate/AMP-PNP/Mg | | Descriptor: | Inositol-trisphosphate 3-kinase A, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
1W7P
 
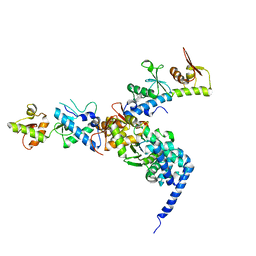 | | The crystal structure of endosomal complex ESCRT-II (VPS22/VPS25/VPS36) | | Descriptor: | VPS22, YPL002C, VPS25, ... | | Authors: | Teo, H, Perisic, O, Gonzalez, B, Williams, R.L. | | Deposit date: | 2004-09-07 | | Release date: | 2004-09-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Escrt-II, an Endosome-Associated Complex Required for Protein Sorting: Crystal Structure and Interactions with Escrt-III and Membranes
Dev.Cell, 7, 2004
|
|
7AUM
 
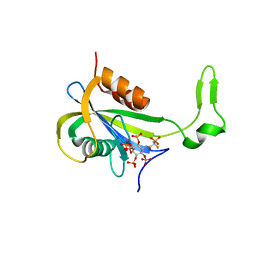 | | Yeast Diphosphoinositol Polyphosphate Phosphohydrolase DDP1 in complex with 5-PCF2Am-InsP5 | | Descriptor: | (1,1-difluoro-2-oxo-2-{[(1s,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]amino}ethyl)phosphonic acid, Diphosphoinositol polyphosphate phosphohydrolase DDP1 | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2020-11-03 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Multiple substrate recognition by yeast diadenosine and diphosphoinositol polyphosphate phosphohydrolase through phosphate clamping.
Sci Adv, 7, 2021
|
|
7AUU
 
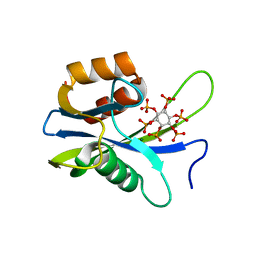 | |
7AUT
 
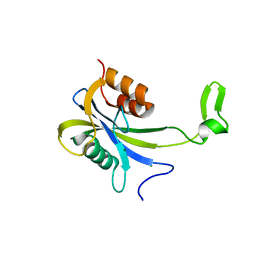 | |
7AUN
 
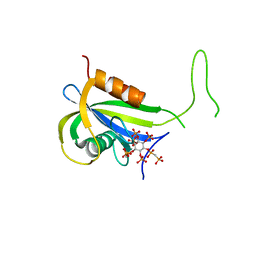 | | Yeast Diphosphoinositol Polyphosphate Phosphohydrolase DDP1 in complex with PCP-InsP8 | | Descriptor: | Diphosphoinositol polyphosphate phosphohydrolase DDP1, MAGNESIUM ION, {[(1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl]bis[oxy(hydroxyphosphoryl)methanediyl]}bis(phosphonic acid) | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2020-11-03 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Multiple substrate recognition by yeast diadenosine and diphosphoinositol polyphosphate phosphohydrolase through phosphate clamping.
Sci Adv, 7, 2021
|
|
7AUP
 
 | | Yeast Diphosphoinositol Polyphosphate Phosphohydrolase DDP1 in complex with PCP-InsP7 | | Descriptor: | Diphosphoinositol polyphosphate phosphohydrolase DDP1, MAGNESIUM ION, [oxidanyl-[(2~{S},3~{S},5~{R},6~{S})-2,3,4,5,6-pentaphosphonooxycyclohexyl]oxy-phosphoryl]methylphosphonic acid | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2020-11-03 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Multiple substrate recognition by yeast diadenosine and diphosphoinositol polyphosphate phosphohydrolase through phosphate clamping.
Sci Adv, 7, 2021
|
|
