7E09
 
 | |
7E06
 
 | | Trans-3/4-proline-hydroxylase H11 with AKG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.G, Yang, L.Y. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Trans-3/4-proline-hydroxylase H11 with AKG and L-proline
To Be Published
|
|
7E08
 
 | |
3ZEE
 
 | | Electron cyro-microscopy helical reconstruction of Par-3 N terminal domain | | Descriptor: | PARTITIONING DEFECTIVE 3 HOMOLOG | | Authors: | Zhang, Y, Wang, W, Chen, J, Zhang, K, Gao, F, Gong, W, Zhang, M, Sun, F, Feng, W. | | Deposit date: | 2012-12-05 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural Insights Into the Intrinsic Self-Assembly of Par-3 N-Terminal Domain.
Structure, 21, 2013
|
|
1SZZ
 
 | | Crystal structure of peptide deformylase from Leptospira Interrogans complexed with inhibitor actinonin | | Descriptor: | ACTINONIN, Peptide deformylase, ZINC ION | | Authors: | Zhou, Z, Song, X, Li, Y, Gong, W. | | Deposit date: | 2004-04-06 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Novel conformational states of peptide deformylase from pathogenic bacterium Leptospira interrogans: implications for population shift
J.Biol.Chem., 280, 2005
|
|
1T8P
 
 | | Crystal structure of Human erythrocyte 2,3-bisphosphoglycerate mutase | | Descriptor: | Bisphosphoglycerate mutase | | Authors: | Wang, Y, Wei, Z, Bian, Q, Cheng, Z, Wan, M, Liu, L, Gong, W. | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human bisphosphoglycerate mutase
J.Biol.Chem., 279, 2004
|
|
1T2L
 
 | | Three Crystal Structures of Human Coactosin-like Protein | | Descriptor: | Coactosin-like protein | | Authors: | Liu, L, Wei, Z, Chen, Z, Wang, Y, Gong, W. | | Deposit date: | 2004-04-22 | | Release date: | 2004-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Coactosin-like Protein
J.Mol.Biol., 344, 2004
|
|
1SV2
 
 | | Crystal Structure of Peptide Deformylase from Leptospira Interrogans (LiPDF) at pH7.5 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, FORMIC ACID, Peptide deformylase, ... | | Authors: | Zhou, Z, Song, X, Li, Y, Gong, W. | | Deposit date: | 2004-03-27 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel conformational states of peptide deformylase from pathogenic bacterium Leptospira interrogans: implications for population shift
J.Biol.Chem., 280, 2005
|
|
4U7U
 
 | | Crystal structure of RNA-guided immune Cascade complex from E.coli | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Zhao, H, Sheng, G, Wang, J, Wang, M, Bunkoczi, G, Gong, W, Wei, Z, Wang, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli
Nature, 515, 2014
|
|
3F5O
 
 | | Crystal Structure of hTHEM2(undecan-2-one-CoA) complex | | Descriptor: | CHLORIDE ION, COENZYME A, HEXAETHYLENE GLYCOL, ... | | Authors: | Xu, H, Gong, W. | | Deposit date: | 2008-11-04 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mechanisms of human hotdog-fold thioesterase 2 (hTHEM2) substrate recognition and catalysis illuminated by a structure and function based analysis
Biochemistry, 48, 2009
|
|
4RSL
 
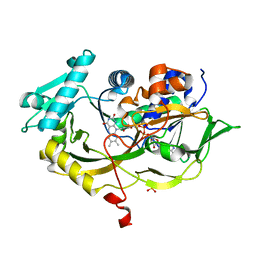 | | Structure of fructosyl peptide oxidase from E. terrenum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl peptide oxidase, PHOSPHATE ION | | Authors: | Gan, W, Gao, F, Xing, K, Jia, M, Liu, H, Gong, W. | | Deposit date: | 2014-11-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the substrate specificity of the FPOD/FAOD family revealed by fructosyl peptide oxidase from Eupenicillium terrenum
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6D26
 
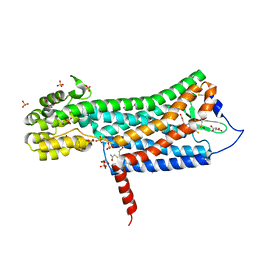 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with fevipiprant | | Descriptor: | OLEIC ACID, Prostaglandin D2 receptor 2, Endolysin chimera, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
6D27
 
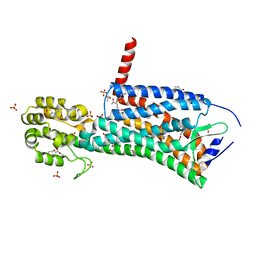 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with CAY10471 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, OLEIC ACID, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
1TXC
 
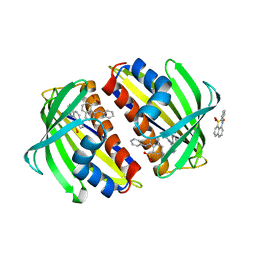 | |
1TW0
 
 | |
1VEY
 
 | | Crystal Structure of Peptide Deformylase from Leptospira Interrogans (LiPDF) at pH7.0 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Peptide deformylase, ZINC ION | | Authors: | Zhou, Z, Song, X, Li, Y, Gong, W. | | Deposit date: | 2004-04-06 | | Release date: | 2005-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Novel conformational states of peptide deformylase from pathogenic bacterium Leptospira interrogans: implications for population shift
J.Biol.Chem., 280, 2005
|
|
1VEZ
 
 | | Crystal Structure of Peptide Deformylase from Leptospira Interrogans(LiPDF) at pH8.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, Peptide deformylase, ... | | Authors: | Zhou, Z, Song, X, Li, Y, Gong, W. | | Deposit date: | 2004-04-06 | | Release date: | 2005-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel conformational states of peptide deformylase from pathogenic bacterium Leptospira interrogans: implications for population shift
J.Biol.Chem., 280, 2005
|
|
1VEV
 
 | | Crystal structure of peptide deformylase from Leptospira Interrogans (LiPDF) at pH6.5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FORMIC ACID, Peptide deformylase, ... | | Authors: | Zhou, Z, Song, X, Li, Y, Gong, W. | | Deposit date: | 2004-04-06 | | Release date: | 2005-08-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Novel conformational states of peptide deformylase from pathogenic bacterium Leptospira interrogans: implications for population shift
J.Biol.Chem., 280, 2005
|
|
4RVZ
 
 | | Crystal structure of tRNA fluorescent labeling enzyme | | Descriptor: | MAGNESIUM ION, N-(4-aminobutyl)-2-azidoacetamide, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Dong, J, Li, F, Wang, J, Gong, W. | | Deposit date: | 2014-11-29 | | Release date: | 2015-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A covalent approach for site-specific RNA labeling in Mammalian cells.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4DZD
 
 | |
1HUZ
 
 | | CRYSTAL STRUCTURE OF DNA POLYMERASE COMPLEXED WITH DNA AND CR-PCP | | Descriptor: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*CP*T)-3', CHROMIUM ION, ... | | Authors: | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | Deposit date: | 2001-01-04 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
1HUO
 
 | | CRYSTAL STRUCTURE OF DNA POLYMERASE BETA COMPLEXED WITH DNA AND CR-TMPPCP | | Descriptor: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*C)-3', CHROMIUM ION, ... | | Authors: | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | Deposit date: | 2001-01-04 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
3K3N
 
 | | Crystal structure of the catalytic core domain of human PHF8 | | Descriptor: | FE (II) ION, PHD finger protein 8 | | Authors: | Yu, L, Wang, Y, Huang, S, Wang, J, Deng, Z, Wu, W, Gong, W, Chen, Z. | | Deposit date: | 2009-10-03 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into a novel histone demethylase PHF8
Cell Res., 20, 2010
|
|
3K3O
 
 | | Crystal structure of the catalytic core domain of human PHF8 complexed with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, PHD finger protein 8 | | Authors: | Yu, L, Wang, Y, Huang, S, Wang, J, Deng, Z, Wu, W, Gong, W, Chen, Z. | | Deposit date: | 2009-10-03 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into a novel histone demethylase PHF8
Cell Res., 20, 2010
|
|
1L2Q
 
 | | Crystal Structure of the Methanosarcina barkeri Monomethylamine Methyltransferase (MtmB) | | Descriptor: | AMMONIUM ION, monomethylamine methyltransferase | | Authors: | Hao, B, Gong, W, Ferguson, T.K, James, C.M, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2002-02-24 | | Release date: | 2002-06-05 | | Last modified: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A new UAG-encoded residue in the structure of a methanogen methyltransferase.
Science, 296, 2002
|
|
