1I74
 
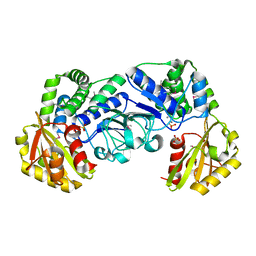 | | STREPTOCOCCUS MUTANS INORGANIC PYROPHOSPHATASE | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, PROBABLE MANGANESE-DEPENDENT INORGANIC PYROPHOSPHATASE, ... | | Authors: | Merckel, M.C, Fabrichniy, I.P, Goldman, A, Lahti, R, Salminen, A. | | Deposit date: | 2001-03-07 | | Release date: | 2001-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Streptococcus mutans pyrophosphatase: a new fold for an old mechanism.
Structure, 9, 2001
|
|
1MUC
 
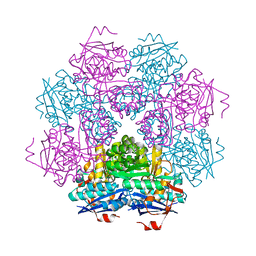 | | STRUCTURE OF MUCONATE LACTONIZING ENZYME AT 1.85 ANGSTROMS RESOLUTION | | Descriptor: | MANGANESE (II) ION, MUCONATE LACTONIZING ENZYME | | Authors: | Helin, S, Kahn, P.C, Guha, B.H.L, Mallows, D.J, Goldman, A. | | Deposit date: | 1995-09-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The refined X-ray structure of muconate lactonizing enzyme from Pseudomonas putida PRS2000 at 1.85 A resolution.
J.Mol.Biol., 254, 1995
|
|
3DAW
 
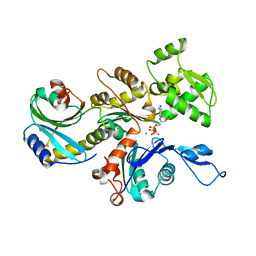 | | Structure of the actin-depolymerizing factor homology domain in complex with actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Paavilainen, V.O, Oksanen, E, Goldman, A, Lappalainen, P. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the actin-depolymerizing factor homology domain in complex with actin
J.Cell Biol., 182, 2008
|
|
2M4F
 
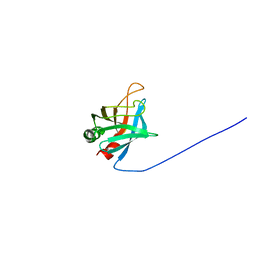 | | Solution Structure of Outer surface protein E | | Descriptor: | Outer surface protein E | | Authors: | Bhattacharjee, A, Oeemig, J.S, Kolodziejczyk, R, Meri, T, Kajander, T, Iwai, H, Jokiranta, T, Goldman, A. | | Deposit date: | 2013-02-05 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Complement Evasion by Lyme Disease Pathogen Borrelia burgdorferi
J.Biol.Chem., 288, 2013
|
|
4AV6
 
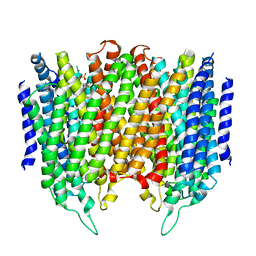 | | Crystal structure of Thermotoga maritima sodium pumping membrane integral pyrophosphatase at 4 A in complex with phosphate and magnesium | | Descriptor: | K(+)-STIMULATED PYROPHOSPHATE-ENERGIZED SODIUM PUMP, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kajander, T, Kellosalo, J, Kogan, K, Pokharel, K, Goldman, A. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Structure and Catalytic Cycle of a Sodium-Pumping Pyrophosphatase.
Science, 337, 2012
|
|
3FUB
 
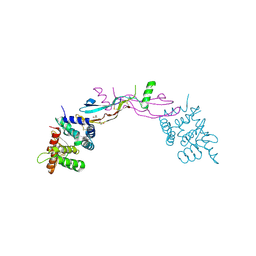 | | Crystal structure of GDNF-GFRalpha1 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Parkash, V, Goldman, A. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Comparison of GFL-GFRalpha complexes: further evidence relating GFL bend angle to RET signalling
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1FAJ
 
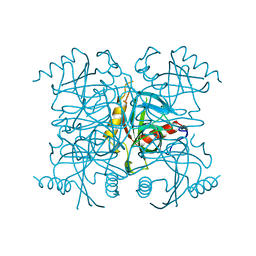 | | INORGANIC PYROPHOSPHATASE | | Descriptor: | SOLUBLE INORGANIC PYROPHOSPHATASE | | Authors: | Kankare, J.A, Salminen, T, Goldman, A. | | Deposit date: | 1996-01-10 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Escherichia coli inorganic pyrophosphatase at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4BWC
 
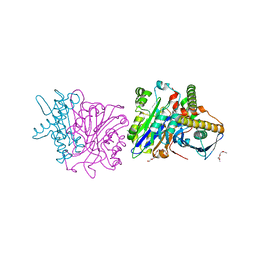 | | X-ray structure of a phospholiapse B like protein 1 from bovine kidneys | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Repo, H, Kuokkanen, E, Oksanen, E, Goldman, A, Heikinheimo, P. | | Deposit date: | 2013-07-01 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Is the Bovine Lysosomal Phospholipase B-Like Protein an Amidase?
Proteins, 82, 2014
|
|
4AV3
 
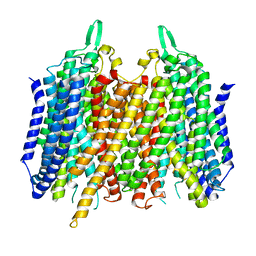 | | Crystal structure of Thermotoga Maritima sodium pumping membrane integral pyrophosphatase with metal ions in active site | | Descriptor: | CALCIUM ION, K(+)-STIMULATED PYROPHOSPHATE-ENERGIZED SODIUM PUMP, MAGNESIUM ION | | Authors: | Kajander, T, Kogan, K, Kellosalo, J, Pokharel, K, Goldman, A. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure and Catalytic Cycle of a Sodium-Pumping Pyrophosphatase.
Science, 337, 2012
|
|
1HIB
 
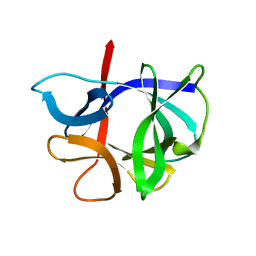 | | THE STRUCTURE OF AN INTERLEUKIN-1 BETA MUTANT WITH REDUCED BIOACTIVITY SHOWS MULTIPLE SUBTLE CHANGES IN CONFORMATION THAT AFFECT PROTEIN-PROTEIN RECOGNITION | | Descriptor: | INTERLEUKIN-1 BETA | | Authors: | Camacho, N.P, Smith, D.R, Goldman, A, Schneider, B, Green, D, Young, P.R, Berman, H.M. | | Deposit date: | 1993-03-29 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of an interleukin-1 beta mutant with reduced bioactivity shows multiple subtle changes in conformation that affect protein-protein recognition.
Biochemistry, 32, 1993
|
|
2XQH
 
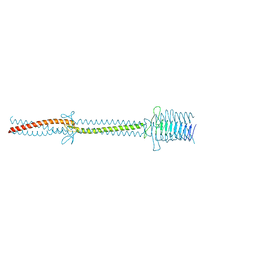 | | Crystal structure of an immunoglobulin-binding fragment of the trimeric autotransporter adhesin EibD | | Descriptor: | CHLORIDE ION, IMMUNOGLOBULIN-BINDING PROTEIN EIBD | | Authors: | Leo, J.C, Lyskowski, A, Hartmann, M, Schwarz, H, Linke, D, Lupas, A.N, Goldman, A. | | Deposit date: | 2010-09-02 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Structure of E. Coli Igg-Binding Protein D Suggests a General Model for Bending and Binding in Trimeric Autotransporter Adhesins.
Structure, 19, 2011
|
|
2PK0
 
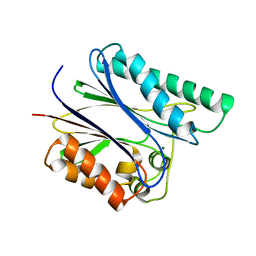 | | Structure of the S. agalactiae serine/threonine phosphatase at 2.65 resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rantanen, M.K, Lehtio, L, Rajagopal, L, Rubens, C.E, Goldman, A. | | Deposit date: | 2007-04-17 | | Release date: | 2007-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of Streptococcus agalactiae serine/threonine phosphatase. The subdomain conformation is coupled to the binding of a third metal ion
Febs J., 274, 2007
|
|
117E
 
 | | THE R78K AND D117E ACTIVE SITE VARIANTS OF SACCHAROMYCES CEREVISIAE SOLUBLE INORGANIC PYROPHOSPHATASE: STRUCTURAL STUDIES AND MECHANISTIC IMPLICATIONS | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Tuominen, V, Heikinheimo, P, Kajander, T, Torkkel, T, Hyytia, T, Kapyla, J, Lahti, R, Cooperman, B.S, Goldman, A. | | Deposit date: | 1998-09-15 | | Release date: | 1998-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The R78K and D117E active-site variants of Saccharomyces cerevisiae soluble inorganic pyrophosphatase: structural studies and mechanistic implications.
J.Mol.Biol., 284, 1998
|
|
1WGI
 
 | |
1WGJ
 
 | |
5ML9
 
 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer F4, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer F4 with specificity for Fc gamma receptor IIIa, CHLORIDE ION, ... | | Authors: | Robinson, J.I, Tomlinson, D.C, Baxter, E.W, Owen, R.L, Thomsen, M, Win, S.J, Nettleship, J.E, Tiede, C, Foster, R.J, Waterhouse, M.P, Harris, S.A, Owens, R.J, Fishwick, C.W.G, Goldman, A, McPherson, M.J, Morgan, A.W. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5MN2
 
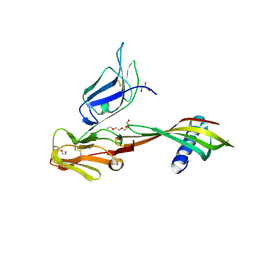 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer G3, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer G3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Robinson, J.I, Owen, R.L, Tomlinson, D.C, Baxter, E.W, Nettleship, J.E, Waterhouse, M.P, Harris, S.A, Owens, R.J, McPherson, M.J, Morgan, A.W, Tiede, C, Goldman, A, Thomsen, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5LZQ
 
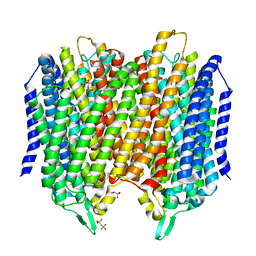 | | Crystal structure of Thermotoga maritima sodium pumping membrane integral pyrophosphatase in complex with imidodiphosphate and magnesium, and with bound sodium ion | | Descriptor: | IMIDODIPHOSPHORIC ACID, K(+)-stimulated pyrophosphate-energized sodium pump, MAGNESIUM ION, ... | | Authors: | Wilkinson, C, Kellosalo, J, Kajander, T, Goldman, A. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Membrane pyrophosphatases from Thermotoga maritima and Vigna radiata suggest a conserved coupling mechanism.
Nat Commun, 7, 2016
|
|
1F9C
 
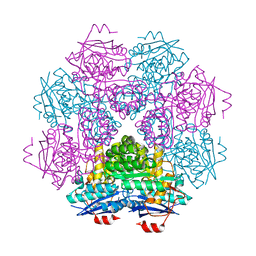 | | CRYSTAL STRUCTURE OF MLE D178N VARIANT | | Descriptor: | MANGANESE (II) ION, PROTEIN (MUCONATE CYCLOISOMERASE I) | | Authors: | Kajander, T, Lehtio, L, Kahn, P.C, Goldman, A. | | Deposit date: | 2000-07-10 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Buried charged surface in proteins.
Structure Fold.Des., 8, 2000
|
|
5LZR
 
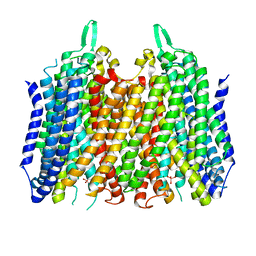 | | Crystal structure of Thermotoga maritima sodium pumping membrane integral pyrophosphatase in complex with tungstate and magnesium | | Descriptor: | K(+)-stimulated pyrophosphate-energized sodium pump, MAGNESIUM ION, TUNGSTATE(VI)ION | | Authors: | Wilkinson, C, Kellosalo, J, Kajander, T, Goldman, A. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Membrane pyrophosphatases from Thermotoga maritima and Vigna radiata suggest a conserved coupling mechanism.
Nat Commun, 7, 2016
|
|
8CHA
 
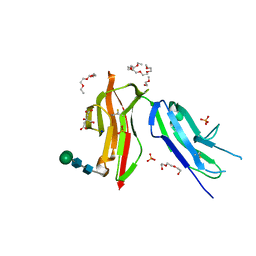 | | Fc gamma RIIa 27W/131H variant ectodomain | | Descriptor: | 1,2-DIMETHOXYETHANE, 2-[2-(2-ethoxyethoxy)ethoxy]ethanol, 3,6,9,12,15,18-HEXAOXAICOSANE, ... | | Authors: | Foy, E.G, Thomsen, M, Goldman, A, Robinson, J.I. | | Deposit date: | 2023-02-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fc gamma RIIa 27W/131H variant ectodomain
To Be Published
|
|
1Q8D
 
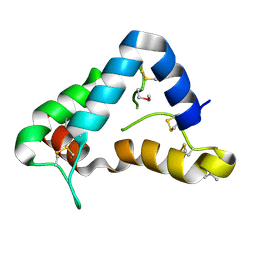 | | The crystal structure of GDNF family co-receptor alpha 1 domain 3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GDNF family receptor alpha 1 | | Authors: | Leppanen, V.M, Bespalov, M.M, Runeberg-Roos, P, Puurand, U, Merits, A, Saarma, M, Goldman, A. | | Deposit date: | 2003-08-21 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of GFRalpha1 domain 3 reveals new insights into GDNF binding and RET activation.
Embo J., 23, 2004
|
|
1E9G
 
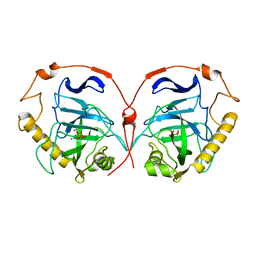 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, MANGANESE (II) ION, PHOSPHATE ION | | Authors: | Heikinheimo, P, Tuominen, V, Ahonen, A.-K, Teplyakov, A, Cooperman, B.S, Baykov, A.A, Lahti, R, Goldman, A. | | Deposit date: | 2000-10-12 | | Release date: | 2001-03-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Toward a Quantum-Mechanical Description of Metal-Assisted Phosphoryl Transfer in Pyrophosphatase
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1E6A
 
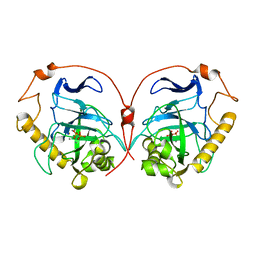 | | Fluoride-inhibited substrate complex of Saccharomyces cerevisiae inorganic pyrophosphatase | | Descriptor: | FLUORIDE ION, INORGANIC PYROPHOSPHATASE, MANGANESE (II) ION, ... | | Authors: | Heikinheimo, P, Tuominen, V, Ahonen, A.-K, Teplyakov, A, Cooperman, B.S, Baykov, A.A, Lahti, R, Goldman, A. | | Deposit date: | 2000-08-09 | | Release date: | 2001-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Toward a quantum-mechanical description of metal-assisted phosphoryl transfer in pyrophosphatase.
Proc. Natl. Acad. Sci. U.S.A., 98, 2001
|
|
2AWU
 
 | | Synapse associated protein 97 PDZ2 domain variant C378G | | Descriptor: | AHH, Synapse-associated protein 97 | | Authors: | Von Ossowski, I, Oksanen, E, Von Ossowski, L, Cai, C, Sundberg, M, Goldman, A, Keinanen, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of the second PDZ domain of SAP97 in complex with a GluR-A C-terminal peptide
Febs J., 273, 2006
|
|
