8AB3
 
 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in complex with oxamate, NADH and FBP. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1,6-di-O-phosphono-beta-D-fructofuranose, L-lactate dehydrogenase, ... | | Authors: | Robin, A.Y, Girard, E, Madern, D. | | Deposit date: | 2022-07-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
8AB2
 
 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in its apo form. | | Descriptor: | 1,2-ETHANEDIOL, L-lactate dehydrogenase, TERBIUM(III) ION, ... | | Authors: | Robin, A.Y, Girard, E, Madern, D. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
3EPV
 
 | | X-ray Structure of the Metal-sensor CnrX in both the Apo- and Copper-bound Forms | | Descriptor: | COPPER (II) ION, Nickel and cobalt resistance protein cnrR | | Authors: | Pompidor, G, Maillard, A.P, Girard, E, Gambarelli, S, Kahn, R, Coves, J. | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | X-ray structure of the metal-sensor CnrX in both the apo- and copper-bound forms.
Febs Lett., 2008
|
|
8OWC
 
 | |
7QO8
 
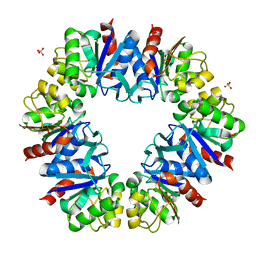 | |
3F2M
 
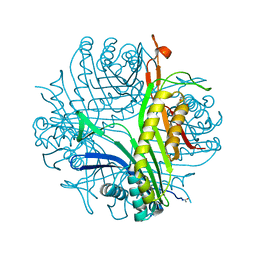 | | Urate oxidase complexed with 8-azaxanthine at 150 MPa | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase | | Authors: | Colloc'h, N, Girard, E, Kahn, R, Fourme, R. | | Deposit date: | 2008-10-30 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function perturbation and dissociation of tetrameric urate oxidase by high hydrostatic pressure.
Biophys.J., 98, 2010
|
|
4BGV
 
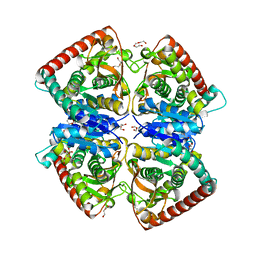 | |
4BGU
 
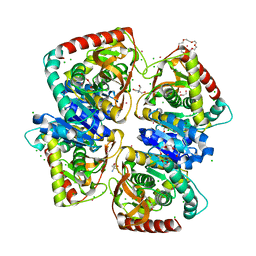 | | 1.50 A resolution structure of the malate dehydrogenase from Haloferax volcanii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Talon, R, Madern, D, Girard, E. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.487 Å) | | Cite: | Insight Into Structural Evolution of Extremophilic Proteins
To be Published
|
|
5NGJ
 
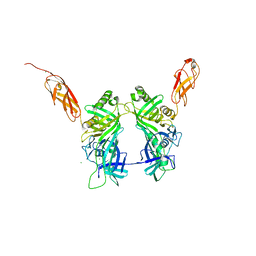 | | Crystal structure of pb6, major tail tube protein of bacteriophage T5 | | Descriptor: | CHLORIDE ION, Tail tube protein | | Authors: | Arnaud, C.-A, Effantin, G, Vives, C, Engilberge, S, Bacia, M, Boulanger, P, Girard, E, Schoehn, G, Breyton, C. | | Deposit date: | 2017-03-17 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacteriophage T5 tail tube structure suggests a trigger mechanism for Siphoviridae DNA ejection.
Nat Commun, 8, 2017
|
|
6Q3T
 
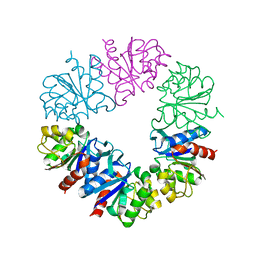 | | Structure of Protease1 from Pyrococcus horikoshii at room temperature in ChipX microfluidic device | | Descriptor: | Deglycase PH1704 | | Authors: | de Wijn, R, Engilberge, S, Olieric, V, Girard, E, Sauter, C. | | Deposit date: | 2018-12-04 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
6QSS
 
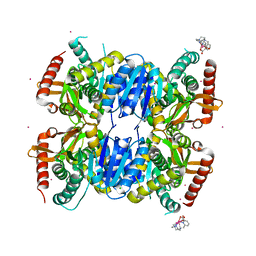 | | Crystal Structure of Ignicoccus islandicus malate dehydrogenase co-crystallized with 10 mM Tb-Xo4 | | Descriptor: | CHLORIDE ION, Malate dehydrogenase, TERBIUM(III) ION, ... | | Authors: | Roche, J, Girard, E, Madern, D. | | Deposit date: | 2019-02-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | SOLUTION SCATTERING (1.892 Å), X-RAY DIFFRACTION | | Cite: | The archaeal LDH-like malate dehydrogenase from Ignicoccus islandicus displays dual substrate recognition, hidden allostery and a non-canonical tetrameric oligomeric organization.
J.Struct.Biol., 208, 2019
|
|
4CL3
 
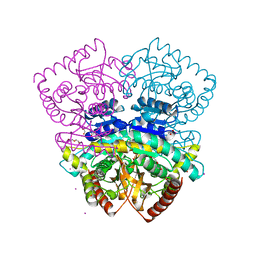 | | 1.70 A resolution structure of the malate dehydrogenase from Chloroflexus aurantiacus | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Talon, R, Madern, D, Girard, E. | | Deposit date: | 2014-01-11 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | An Experimental Point of View on Hydration/Solvation in Halophilic Proteins.
Front.Microbiol., 5, 2014
|
|
8Q8O
 
 | |
8OSO
 
 | | GTPase HRAS in complex with Zn-cyclen under 500 MPa pressure | | Descriptor: | 1,4,7,10-tetraazacyclododecane, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Colloc'h, N, Prange, T, Girard, E, Kalbitzer, H.R. | | Deposit date: | 2023-04-19 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High Pressure Promotes Binding of the Allosteric Inhibitor Zn 2+ -Cyclen in Crystals of Activated H-Ras.
Chemistry, 30, 2024
|
|
8OSM
 
 | | GTPASE HRAS IN COMPLEX WITH ZN-CYCLEN AT 200 MPA PRESSURE | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Colloc'h, N, Girard, E, Prange, T, Kalbitzer, H.R. | | Deposit date: | 2023-04-19 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High Pressure Promotes Binding of the Allosteric Inhibitor Zn 2+ -Cyclen in Crystals of Activated H-Ras.
Chemistry, 30, 2024
|
|
8OSN
 
 | | GTPASE HRAS IN COMPLEX WITH ZN-CYCLEN AT AMBIENT PRESSURE | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Colloc'h, N, Prange, T, Girard, E, Kalbitzer, H.R. | | Deposit date: | 2023-04-19 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High Pressure Promotes Binding of the Allosteric Inhibitor Zn 2+ -Cyclen in Crystals of Activated H-Ras.
Chemistry, 30, 2024
|
|
2PLB
 
 | | D(GTATACC) under hydrostatic pressure of 1.39 GPa | | Descriptor: | 5'-D(*DGP*DGP*DTP*DAP*DTP*DAP*DCP*DC)-3', SPERMINE | | Authors: | Prange, T, Girard, E, Kahn, R, Fourme, R. | | Deposit date: | 2007-04-19 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Adaptation of the base-paired double-helix molecular architecture to extreme pressure.
Nucleic Acids Res., 35, 2007
|
|
2PL4
 
 | |
8RWL
 
 | | Crystal structure of Methanopyrus kandleri malate dehydrogenase mutant 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Malate dehydrogenase, ... | | Authors: | Coquille, S, Roche, J, Engilberge, S, Girard, E, Madern, D. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Navigating the conformational landscape of an enzyme. Stabilization of a low populated conformer by evolutionary mutations triggers Allostery into a non-allosteric enzyme.
To Be Published
|
|
2QMI
 
 | | Structure of the octameric penicillin-binding protein homologue from Pyrococcus abyssi | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, LUTETIUM (III) ION, Pbp related beta-lactamase | | Authors: | Delfosse, V, Girard, E, Moulinier, L, Schultz, P, Mayer, C. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the archaeal pab87 peptidase reveals a novel self-compartmentalizing protease family
Plos One, 4, 2009
|
|
2Y39
 
 | | Ni-bound form of Cupriavidus metallidurans CH34 CnrXs | | Descriptor: | ACETATE ION, NICKEL (II) ION, NICKEL AND COBALT RESISTANCE PROTEIN CNRR | | Authors: | Trepreau, J, Girard, E, Maillard, A.P, de Rosny, E, Petit-Haertlein, I, Kahn, R, Coves, J. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural Basis for Metal Sensing by Cnrx.
J.Mol.Biol., 408, 2011
|
|
2Y3D
 
 | | Zn-bound form of Cupriavidus metallidurans CH34 CnrXs | | Descriptor: | CHLORIDE ION, NICKEL AND COBALT RESISTANCE PROTEIN CNRR, ZINC ION | | Authors: | Trepreau, J, Girard, E, Maillard, A.P, de Rosny, E, Petit-Haertlein, I, Kahn, R, Coves, J. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Metal Sensing by Cnrx.
J.Mol.Biol., 408, 2011
|
|
2Y3G
 
 | | Se-Met form of Cupriavidus metallidurans CH34 CnrXs | | Descriptor: | CHLORIDE ION, GLYCEROL, NICKEL AND COBALT RESISTANCE PROTEIN CNRR, ... | | Authors: | Trepreau, J, Girard, E, Maillard, A.P, de Rosny, E, Petit-Haertlein, I, Kahn, R, Coves, J. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Basis for Metal Sensing by Cnrx.
J.Mol.Biol., 408, 2011
|
|
5LDA
 
 | | Structure of deubiquitinating enzyme homolog (Pyrococcus furiosus JAMM1) in complex with ubiquitin-like SAMP2. | | Descriptor: | GLYCEROL, JAMM1, SAMP2, ... | | Authors: | Cao, S, Engilberge, S, Girard, E, Gabel, F, Franzetti, B, Maupin-Furlow, J.A. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-21 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight into Ubiquitin-Like Protein Recognition and Oligomeric States of JAMM/MPN(+) Proteases.
Structure, 25, 2017
|
|
5LD9
 
 | | Structure of deubiquitinating enzyme homolog, Pyrococcus furiosus JAMM1. | | Descriptor: | CHLORIDE ION, JAMM1, ZINC ION | | Authors: | Maupin-Furlow, J.A, Franzetti, B, Cao, S, Girard, E, Gabel, F, Engilberge, S. | | Deposit date: | 2016-06-24 | | Release date: | 2017-05-17 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Structural Insight into Ubiquitin-Like Protein Recognition and Oligomeric States of JAMM/MPN(+) Proteases.
Structure, 25, 2017
|
|
