4YP2
 
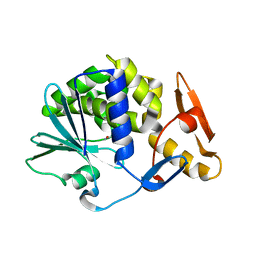 | | Cleavage of nicotinamide adenine dinucleotides by the ribosome inactivating protein from Momordica charantia | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NICOTINAMIDE, Ribosome-inactivating protein momordin I | | Authors: | Vinkovic, M, Hussain, J, Wood, G.E, Gill, R, Wood, S.P. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Cleavage of nicotinamide adenine dinucleotide by the ribosome-inactivating protein from Momordica charantia.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3EQ1
 
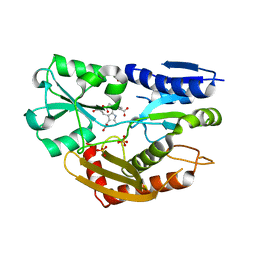 | | The Crystal Structure of Human Porphobilinogen Deaminase at 2.8A resolution | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase, SULFATE ION | | Authors: | Kolstoe, S.E, Gill, R, Mohammed, F, Wood, S.P. | | Deposit date: | 2008-09-30 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human porphobilinogen deaminase at 2.8 A: the molecular basis of acute intermittent porphyria
Biochem.J., 420, 2009
|
|
1QN2
 
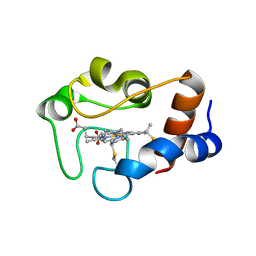 | | cytochrome cH from Methylobacterium extorquens | | Descriptor: | CYTOCHROME CH, HEME C | | Authors: | Read, J, Gill, R, Dales, S.L, Cooper, J.B, Wood, S.P, Anthony, C. | | Deposit date: | 1999-10-13 | | Release date: | 2000-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Molecular Structure of an Unusual Cytochrome C2 Determined at 2.0A; the Cytochrome cH from Methylobacterium Extorquens
Protein Sci., 8, 1999
|
|
1OEW
 
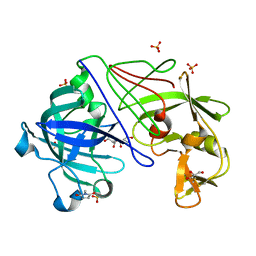 | | ATOMIC RESOLUTION STRUCTURE OF NATIVE ENDOTHIAPEPSIN | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, SERINE, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
1W6S
 
 | | The high resolution structure of methanol dehydrogenase from methylobacterium extorquens | | Descriptor: | CALCIUM ION, GLYCEROL, METHANOL DEHYDROGENASE SUBUNIT 1, ... | | Authors: | Williams, P.A, Coates, L, Mohammed, F, Gill, R, Erskine, P.T, Wood, S.P, Anthony, C, Cooper, J.B. | | Deposit date: | 2004-08-23 | | Release date: | 2004-12-21 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Atomic Resolution Structure of Methanol Dehydrogenase from Methylobacterium Extorquens
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1OEX
 
 | | Atomic Resolution Structure of Endothiapepsin in Complex with a Hydroxyethylene Transition State Analogue Inhibitor H261 | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, INHIBITOR H261, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
1OD1
 
 | | Endothiapepsin PD135,040 complex | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide, SULFATE ION | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Cooper, J.B. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The Structure of Endothiapepsin Complexed with the Gem-Diol Inhibitor Pd-135,040 at 1.37 A
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2IZP
 
 | | BipD - an invasion protein associated with the type-III secretion system of Burkholderia pseudomallei. | | Descriptor: | PUTATIVE MEMBRANE ANTIGEN | | Authors: | Erskine, P.T, Knight, M.J, Ruaux, A, Mikolajek, H, Wong-Fat-Sang, N, Withers, J, Gill, R, Wood, S.P, Wood, M, Fox, G.C, Cooper, J.B. | | Deposit date: | 2006-07-25 | | Release date: | 2006-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High Resolution Structure of Bipd: An Invasion Protein Associated with the Type III Secretion System of Burkholderia Pseudomallei.
J.Mol.Biol., 363, 2006
|
|
1H4J
 
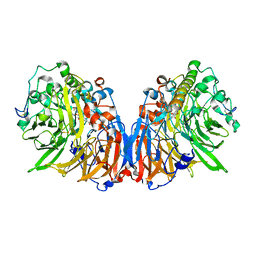 | | Methylobacterium extorquens methanol dehydrogenase D303E mutant | | Descriptor: | CALCIUM ION, Methanol dehydrogenase [cytochrome c] subunit 1, Methanol dehydrogenase [cytochrome c] subunit 2, ... | | Authors: | Mohammed, F, Gill, R, Thompson, D, Cooper, J.B, Wood, S.P, Afolabi, P.R, Anthony, C. | | Deposit date: | 2001-05-11 | | Release date: | 2001-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Site-Directed Mutagenesis and X-Ray Crystallography of the Pqq-Containing Quinoprotein Methanol Dehydrogenase and its Electron Acceptor, Cytochrome C(L)(,)
Biochemistry, 40, 2001
|
|
1W31
 
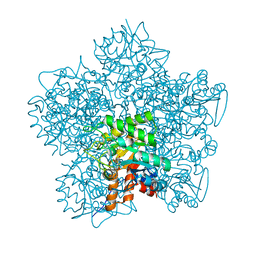 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE 5-HYDROXYLAEVULINIC ACID COMPLEX | | Descriptor: | 5-HYDROXYLAEVULINIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Stauffer, F, Beaven, G.D.E, Gill, R, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2004-07-11 | | Release date: | 2005-08-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with the Inhibitor 5-Hydroxylaevulinic Acid
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1W1Z
 
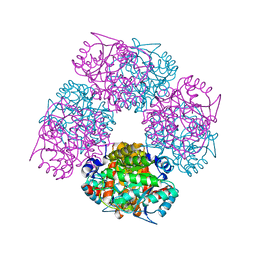 | | Structure of the plant like 5-Aminolaevulinic Acid Dehydratase from Chlorobium vibrioforme | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, LAEVULINIC ACID, MAGNESIUM ION | | Authors: | Coates, L, Beaven, G, Erskine, P.T, Beale, S.I, Avissar, Y.J, Gill, R, Mohammed, F, Wood, S.P, Shoolingin-Jordan, P, Cooper, J.B. | | Deposit date: | 2004-06-24 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The X-ray structure of the plant like 5-aminolaevulinic acid dehydratase from Chlorobium vibrioforme complexed with the inhibitor laevulinic acid at 2.6 A resolution.
J. Mol. Biol., 342, 2004
|
|
1B9G
 
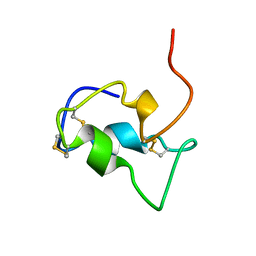 | | INSULIN-LIKE-GROWTH-FACTOR-1 | | Descriptor: | PROTEIN (GROWTH FACTOR IGF-1) | | Authors: | De Wolf, E, Gill, R, Geddes, S, Pitts, J, Wollmer, A, Grotzinger, J. | | Deposit date: | 1999-02-11 | | Release date: | 1999-02-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a mini IGF-1.
Protein Sci., 5, 1996
|
|
4V8Q
 
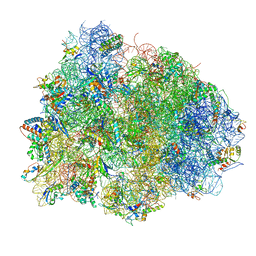 | | Complex of SmpB, a tmRNA fragment and EF-Tu-GDP-Kirromycin with the 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Neubauer, C, Gillet, R, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2011-12-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Decoding in the absence of a codon by tmRNA and SmpB in the ribosome.
Science, 335, 2012
|
|
1QJV
 
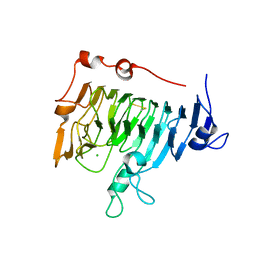 | | Pectin methylesterase PemA from Erwinia chrysanthemi | | Descriptor: | CHLORIDE ION, PECTIN METHYLESTERASE | | Authors: | Jenkins, J, Mayans, O, Smith, D, Worboys, K, Pickersgill, R. | | Deposit date: | 1999-07-05 | | Release date: | 2000-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Three-Dimensional Structure of Erwinia Chrysanthemi Pectin Methylesterase Reveals a Novel Esterase Active Site
J.Mol.Biol., 305, 2001
|
|
3L3P
 
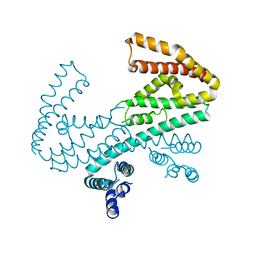 | | Crystal structure of the C-terminal domain of Shigella type III effector IpaH9.8, with a novel domain swap | | Descriptor: | IpaH9.8 | | Authors: | Seyedarabi, A, Sullivan, J.A, Sasakawa, C, Pickersgill, R.W. | | Deposit date: | 2009-12-17 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A disulfide driven domain swap switches off the activity of Shigella IpaH9.8 E3 ligase
Febs Lett., 584, 2010
|
|
3JBH
 
 | | TWO HEAVY MEROMYOSIN INTERACTING-HEADS MOTIFS FLEXIBLE DOCKED INTO TARANTULA THICK FILAMENT 3D-MAP ALLOWS IN DEPTH STUDY OF INTRA- AND INTERMOLECULAR INTERACTIONS | | Descriptor: | MYOSIN 2 ESSENTIAL LIGHT CHAIN STRIATED MUSCLE, MYOSIN 2 HEAVY CHAIN STRIATED MUSCLE, MYOSIN 2 REGULATORY LIGHT CHAIN STRIATED MUSCLE | | Authors: | Alamo, L, Qi, D, Wriggers, W, Pinto, A, Zhu, J, Bilbao, A, Gillilan, R.E, Hu, S, Padron, R. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis.
J. Mol. Biol., 428, 2016
|
|
8A3L
 
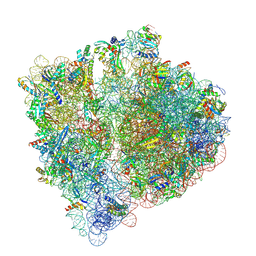 | | Structural insights into the binding of bS1 to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | D'Urso, G, Chat, S, Gillet, R, Giudice, E. | | Deposit date: | 2022-06-08 | | Release date: | 2023-05-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural insights into the binding of bS1 to the ribosome.
Nucleic Acids Res., 51, 2023
|
|
7UXE
 
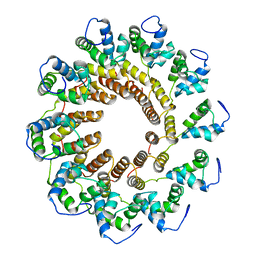 | | Pseudomonas phage E217 small terminase (TerS) | | Descriptor: | Small terminase | | Authors: | Lokareddy, R.K, Hou, C.-F.D, Doll, S.G, Li, F, Gillilan, R, Forti, F, Briani, F, Cingolani, G. | | Deposit date: | 2022-05-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Terminase Subunits from the Pseudomonas-Phage E217.
J.Mol.Biol., 434, 2022
|
|
4X7G
 
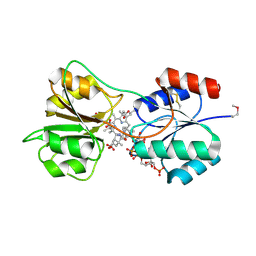 | | CobK precorrin-6A reductase | | Descriptor: | 3-[(1R,2S,3S,7S,11S,16S,17R,18R,19R)-2,7,12,18-tetrakis(2-hydroxy-2-oxoethyl)-3,13-bis(3-hydroxy-3-oxopropyl)-1,2,5,7,11,17-hexamethyl-17-(3-oxidanyl-3-oxidanylidene-prop-1-enyl)-3,6,8,10,15,16,18,19,21,24-decahydrocorrin-8-yl]propanoic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Precorrin-6A reductase | | Authors: | Gu, S, Deery, E, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2014-12-09 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal structure of CobK reveals strand-swapping between Rossmann-fold domains and molecular basis of the reduced precorrin product trap.
Sci Rep, 5, 2015
|
|
9FCT
 
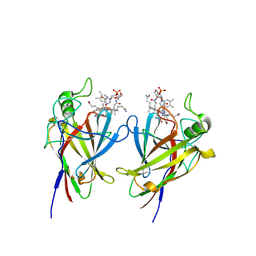 | |
4WFW
 
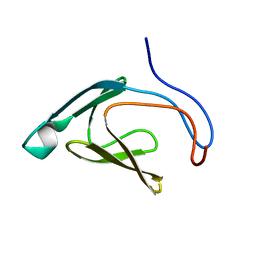 | |
6YOA
 
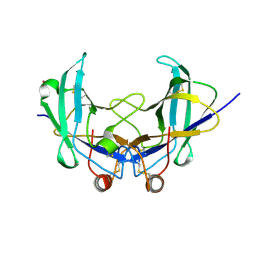 | | Lig v 1 structure and the inflammatory response to the Ole e 1 protein family | | Descriptor: | Major pollen allergen Lig v 1, NICKEL (II) ION | | Authors: | Robledo-Retana, T, Bradley-Clark, J, Croll, T, Rose, R, Stagg, A, Villalba, M, Pickersgill, R. | | Deposit date: | 2020-04-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Lig v 1 structure and the inflammatory response to the Ole e 1 protein family.
Allergy, 75, 2020
|
|
7ACJ
 
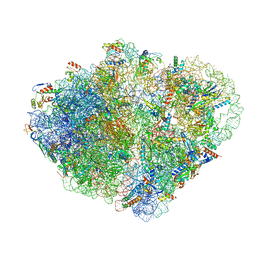 | | Structure of translocated trans-translation complex on E. coli stalled ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Guyomar, C, D'Urso, G, Chat, S, Giudice, E, Gillet, R. | | Deposit date: | 2020-09-11 | | Release date: | 2021-08-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of tmRNA and SmpB as they transit through the ribosome.
Nat Commun, 12, 2021
|
|
7ABZ
 
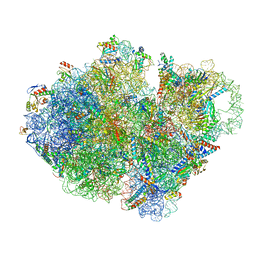 | | Structure of pre-accomodated trans-translation complex on E. coli stalled ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Guyomar, C, D'Urso, G, Chat, S, Giudice, E, Gillet, R. | | Deposit date: | 2020-09-09 | | Release date: | 2021-08-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structures of tmRNA and SmpB as they transit through the ribosome.
Nat Commun, 12, 2021
|
|
7ACR
 
 | | Structure of post-translocated trans-translation complex on E. coli stalled ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Guyomar, C, D'Urso, G, Chat, S, Giudice, E, Gillet, R. | | Deposit date: | 2020-09-11 | | Release date: | 2021-08-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structures of tmRNA and SmpB as they transit through the ribosome.
Nat Commun, 12, 2021
|
|
