8FUI
 
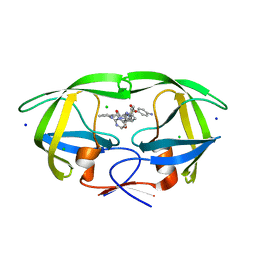 | | HIV-1 wild type protease with GRL-02519A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
4G61
 
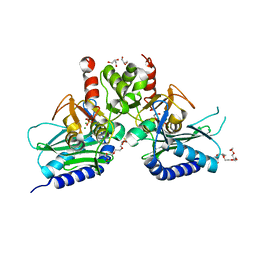 | | Crystal structure of IMPase/NADP phosphatase complexed with Mg2+ and phosphate | | Descriptor: | 1-HYDROXYSULFANYL-4-MERCAPTO-BUTANE-2,3-DIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2012-07-18 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural elucidation of the binding site and mode of inhibition of Li(+) and Mg(2+) in inositol monophosphatase.
Febs J., 281, 2014
|
|
5F24
 
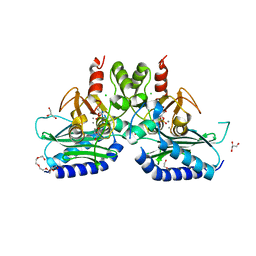 | | Crystal structure of dual specific IMPase/NADP phosphatase bound with D-inositol-1-phosphate | | Descriptor: | CALCIUM ION, CHLORIDE ION, D-MYO-INOSITOL-1-PHOSPHATE, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-12-01 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EYH
 
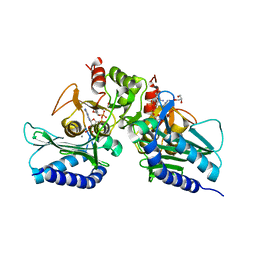 | | Crystal Structure of IMPase/NADP phosphatase complexed with NADP and Ca2+ at pH 7.0 | | Descriptor: | CALCIUM ION, GLYCEROL, Inositol monophosphatase, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-11-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EYG
 
 | | Crystal structure of IMPase/NADP phosphatase complexed with NADP and Ca2+ | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-11-25 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3RYD
 
 | | Crystal structure of Ca bound IMPase family protein from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Inositol monophosphatase family protein, POTASSIUM ION, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2011-05-11 | | Release date: | 2012-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of Staphylococcal dual specific inositol monophosphatase/NADP(H) phosphatase (SAS2203) delineates the molecular basis of substrate specificity
Biochimie, 94, 2012
|
|
3P7X
 
 | | Crystal structure of an atypical two-cysteine peroxiredoxin (SAOUHSC_01822) from Staphylococcus aureus NCTC8325 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Probable thiol peroxidase, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2010-10-13 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of an atypical two-cysteine peroxiredoxin (SAOUHSC_01822) from Staphylococcus aureus NCTC8325
To be Published
|
|
3QMF
 
 | | Crystal strucuture of an inositol monophosphatase family protein (SAS2203) from Staphylococcus aureus MSSA476 | | Descriptor: | Inositol monophosphatase family protein, SULFATE ION | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2011-02-04 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Staphylococcal dual specific inositol monophosphatase/NADP(H) phosphatase (SAS2203) delineates the molecular basis of substrate specificity
Biochimie, 94, 2012
|
|
6A4J
 
 | | Crystal structure of Thioredoxin reductase 2 from Staphylococcus aureus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase | | Authors: | Bose, M, Bhattacharyya, S, Ghosh, A.K, Das, A.K. | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Elucidation of the mechanism of disulfide exchange between staphylococcal thioredoxin2 and thioredoxin reductase2: A structural insight.
Biochimie, 160, 2019
|
|
4NJS
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with non-peptidic inhibitor, GRL008 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-carbamoylphenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
2IDW
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a potent non-peptide inhibitor (UIC-94017) | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Manna, D, Hussain, A.K, Leshchenko, S, Ghosh, A.K, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-09-15 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High Resolution Crystal Structures of HIV-1 Protease with a Potent Non-Peptide Inhibitor (Uic-94017) Active Against Multi-Drug-Resistant Clinical Strains.
J.Mol.Biol., 338, 2004
|
|
4NJT
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Yedidi, R.S, Garimella, H, Chang, S.B, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
6D0E
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-084-13 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-1-(3,5-difluorophenyl)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Central Nervous System (CNS)-Targeting Protease Inhibitors for Drug-Resistant HIV Infection and HIV-Associated CNS Complications.
Antimicrob.Agents Chemother., 63, 2019
|
|
2QCI
 
 | | HIV-1 Protease mutant D30N with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
2QD8
 
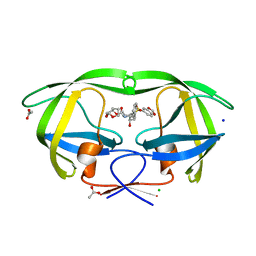 | | HIV-1 Protease Mutant I84V with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
2QD7
 
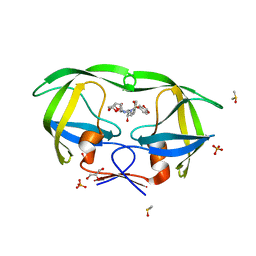 | | HIV-1 Protease Mutant V82A with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
5COK
 
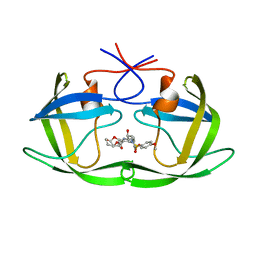 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-0476 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5COP
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-097 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-(4-methoxyphenyl)butan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
2QD6
 
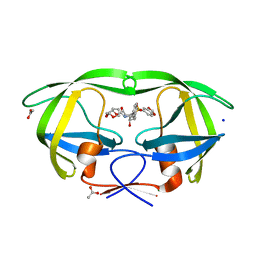 | | HIV-1 Protease Mutant I50V with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
5CON
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-015 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5COO
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-085 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-1-(4-methoxyphenyl)-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
2VKM
 
 | | Crystal structure of GRL-8234 bound to BACE (Beta-secretase) | | Descriptor: | BETA-SECRETASE 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Hong, L, Tang, J, Ghosh, A.K. | | Deposit date: | 2007-12-04 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Potent Memapsin 2 (Beta-Secretase) Inhibitors: Design, Synthesis, Protein-Ligand X-Ray Structure, and in Vivo Evaluation.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2P4J
 
 | | Crystal structure of beta-secretase bond to an inhibitor with Isophthalamide Derivatives at P2-P3 | | Descriptor: | Beta-secretase 1, N-[(1S,2S,4R)-2-HYDROXY-1-ISOBUTYL-5-({(1S)-1-[(ISOPROPYLAMINO)CARBONYL]-2-METHYLPROPYL}AMINO)-4-METHYL-5-OXOPENTYL]-5-[METHYL(METHYLSULFONYL)AMINO]-N'-[(1R)-1-PHENYLETHYL]ISOPHTHALAMIDE | | Authors: | Hong, L, Ghosh, A.K, Tang, J. | | Deposit date: | 2007-03-12 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis, and X-ray structure of potent memapsin 2 (beta-secretase) inhibitors with isophthalamide derivatives as the P2-P3-ligands.
J.Med.Chem., 50, 2007
|
|
2NNP
 
 | | Crystal structure analysis of HIV-1 protease mutant I84V with a inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETIC ACID, GLYCEROL, ... | | Authors: | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
2NMZ
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEASE, SULFATE ION | | Authors: | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-10-23 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
