4HPS
 
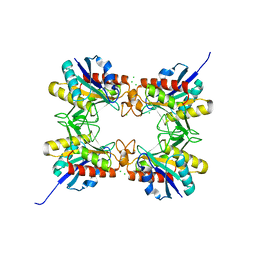 | |
4HKM
 
 | |
4ESO
 
 | | Crystal structure of a putative oxidoreductase protein from Sinorhizobium meliloti 1021 in complex with NADP | | 分子名称: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | 著者 | Ghosh, A, Bhoshle, R, Toro, R, Gizzi, A, Hillerich, B, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-04-23 | | 公開日 | 2012-05-16 | | 実験手法 | X-RAY DIFFRACTION (1.906 Å) | | 主引用文献 | Crystal structure of a putative oxidoreductase protein from Sinorhizobium meliloti 1021 in complex with NADP
To be Published
|
|
3EF0
 
 | |
3EF1
 
 | |
4H15
 
 | |
4GXH
 
 | |
4H16
 
 | |
3RTX
 
 | |
2N6M
 
 | |
2MJX
 
 | | Solution NMR structure of a mismatch DNA | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*TP*AP*CP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*AP*TP*GP*CP*TP*AP*CP*GP*CP*G)-3') | | 著者 | Ghosh, A, Kumar, K.R, Bhunia, A, Chatterjee, S. | | 登録日 | 2014-01-21 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Double GC:GC mismatch in dsDNA enhances local dynamics retaining the DNA footprint: a high-resolution NMR study
Chemmedchem, 9, 2014
|
|
2JR6
 
 | | Solution structure of UPF0434 protein NMA0874. Northeast Structural Genomics Target MR32 | | 分子名称: | UPF0434 protein NMA0874 | | 著者 | Ghosh, A, Singarapu, K.K, Wu, Y, Liu, G, Sukumaran, D, Chen, C.X, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-06-20 | | 公開日 | 2007-07-24 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of UPF0434 protein NMA0874.
To be Published
|
|
4GID
 
 | | Structure of beta-secretase complexed with inhibitor | | 分子名称: | Beta-secretase 1, L-PROLINAMIDE, N-[(2S)-1-({(2S,3R)-3-hydroxy-1-[(2-methylpropyl)amino]-1-oxobutan-2-yl}amino)-3-phenylpropan-2-yl]-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | 著者 | Ghosh, A, Tang, J, Venkateswara, R.K, Yadav, N, Anderson, D, Gavande, N, Huang, X, Terzyan, S. | | 登録日 | 2012-08-08 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-based design of highly selective beta-secretase inhibitors: synthesis, biological evaluation, and protein-ligand X-ray crystal structure.
J.Med.Chem., 55, 2012
|
|
1XS7
 
 | | Crystal Structure of a cycloamide-urethane-derived novel inhibitor bound to human brain memapsin 2 (beta-secretase). | | 分子名称: | Beta-secretase 1, N-[(4S,5S,7R)-8-({(S)-1-[(BENZYLAMINO)OXOMETHYL]-2-METHYLPROPYL}AMINO)-5-HYDROXY-2,7-DIMETHYL-8-OXO-OCT-4-YL]-(4S,7S)-4 -ISOPROPYL-2,5,9-TRIOXO-1-OXA-3,6,10-TRIAZACYCLOHEXADECANE-7-CARBOXAMIDE | | 著者 | Ghosh, A, Devasamudram, T, Hong, L, DeZutter, C, Xu, X, Weerasena, V, Koelsch, G, Bilcer, G, Tang, J. | | 登録日 | 2004-10-18 | | 公開日 | 2004-12-21 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure-based design of cycloamide-urethane-derived novel inhibitors of human brain memapsin 2 (beta-secretase).
Bioorg.Med.Chem.Lett., 15, 2005
|
|
8GAC
 
 | | Crystal structure of a high affinity CTLA-4 binder | | 分子名称: | 1,2-ETHANEDIOL, CTLA-4 binder | | 著者 | Yang, W, Almo, S.C, Baker, D, Ghosh, A. | | 登録日 | 2023-02-22 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Design of High Affinity Binders to Convex Protein Target Sites.
Biorxiv, 2024
|
|
8GAD
 
 | |
8GAB
 
 | | Crystal structure of CTLA-4 in complex with a high affinity CTLA-4 binder | | 分子名称: | CTLA-4 binder, Cytotoxic T-lymphocyte protein 4, POTASSIUM ION | | 著者 | Yang, W, Almo, S.C, Baker, D, Ghosh, A. | | 登録日 | 2023-02-22 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Design of High Affinity Binders to Convex Protein Target Sites.
Biorxiv, 2024
|
|
7RBW
 
 | | Structure of Biliverdin-binding Serpin of Boana punctata (polka-dot tree frog) | | 分子名称: | BILIVERDINE IX ALPHA, Biliverdin bindin serpin | | 著者 | Fedorov, E, Manoilov, K.Y, Verkhusha, V, Almo, S.C, Ghosh, A. | | 登録日 | 2021-07-06 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural and Functional Characterization of a Biliverdin-Binding Near-Infrared Fluorescent Protein From the Serpin Superfamily.
J.Mol.Biol., 434, 2021
|
|
8SWS
 
 | | Structure of K. lactis PNP S42E-H98R variant bound to transition state analog DADMe-IMMUCILLIN G and sulfate | | 分子名称: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | 著者 | Fedorov, E, Ghosh, A. | | 登録日 | 2023-05-19 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Phosphate Binding in PNP Alters Transition-State Analogue Affinity and Subunit Cooperativity.
Biochemistry, 62, 2023
|
|
8SWP
 
 | |
8SWR
 
 | | Structure of K. lactis PNP S42E variant bound to transition state analog DADMe-IMMUCILLIN G and sulfate | | 分子名称: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, GUANINE, Purine nucleoside phosphorylase, ... | | 著者 | Fedorov, E, Ghosh, A. | | 登録日 | 2023-05-19 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Phosphate Binding in PNP Alters Transition-State Analogue Affinity and Subunit Cooperativity.
Biochemistry, 62, 2023
|
|
8SWQ
 
 | | Structure of K. lactis PNP bound to transition state analog DADMe-IMMUCILLIN H and sulfate | | 分子名称: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, GLYCEROL, Purine nucleoside phosphorylase, ... | | 著者 | Fedorov, E, Ghosh, A. | | 登録日 | 2023-05-19 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.979 Å) | | 主引用文献 | Phosphate Binding in PNP Alters Transition-State Analogue Affinity and Subunit Cooperativity.
Biochemistry, 62, 2023
|
|
8SWT
 
 | |
8SWU
 
 | |
9C6V
 
 | | Crystal Structure of a single chain trimer composed of HLA-B*39:06 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 (1 molecule/asymmetric unit) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | 登録日 | 2024-06-09 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 300, 2024
|
|
