3EF1
 
 | |
3RTX
 
 | |
2M2C
 
 | | Solution structure of Duplex DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*AP*GP*CP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*AP*TP*GP*CP*TP*AP*CP*GP*CP*G)-3') | | Authors: | Ghosh, A, Kar, R.K, Chatterjee, S, Bhunia, A. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Indolicidin targets duplex DNA: structural and mechanistic insight through a combination of spectroscopy and microscopy.
Chemmedchem, 9, 2014
|
|
1XS7
 
 | | Crystal Structure of a cycloamide-urethane-derived novel inhibitor bound to human brain memapsin 2 (beta-secretase). | | Descriptor: | Beta-secretase 1, N-[(4S,5S,7R)-8-({(S)-1-[(BENZYLAMINO)OXOMETHYL]-2-METHYLPROPYL}AMINO)-5-HYDROXY-2,7-DIMETHYL-8-OXO-OCT-4-YL]-(4S,7S)-4 -ISOPROPYL-2,5,9-TRIOXO-1-OXA-3,6,10-TRIAZACYCLOHEXADECANE-7-CARBOXAMIDE | | Authors: | Ghosh, A, Devasamudram, T, Hong, L, DeZutter, C, Xu, X, Weerasena, V, Koelsch, G, Bilcer, G, Tang, J. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of cycloamide-urethane-derived novel inhibitors of human brain memapsin 2 (beta-secretase).
Bioorg.Med.Chem.Lett., 15, 2005
|
|
5XDJ
 
 | | Esculentin-1a(1-21)NH2 | | Descriptor: | Esculentin-1A | | Authors: | Ghosh, A, Bhunia, A. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Membrane perturbing activities and structural properties of the frog-skin derived peptide Esculentin-1a(1-21)NH2 and its Diastereomer Esc(1-21)-1c: Correlation with their antipseudomonal and cytotoxic activity
Biochim. Biophys. Acta, 1859, 2017
|
|
5XER
 
 | | TK9 NMR structure in DPC micelle | | Descriptor: | THR-VAL-TYR-VAL-TYR-SER-ARG-VAL-LYS | | Authors: | Ghosh, A, Bhunia, A. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights of a self-assembling 9-residue peptide from the C-terminal tail of the SARS corona virus E-protein in DPC and SDS micelles: A combined high and low resolution spectroscopic study.
Biochim Biophys Acta Biomembr, 1860, 2018
|
|
5DQC
 
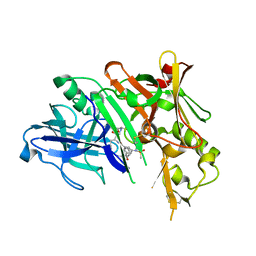 | | Co-crystal of BACE1 with compound 0211 | | Descriptor: | Beta-secretase 1, N-[(2S,3R)-3-hydroxy-4-({(2S,3S)-3-hydroxy-1-[(2-methylpropyl)amino]-1-oxobutan-2-yl}amino)-1-phenylbutan-2-yl]-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Ghosh, A.K, Bhavanam, S.R, Yen, T.-C, Cardenas, E.L, Rao, K.V, Downs, D, Huang, X, Tang, J, Mescar, A.D. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-17 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.4651 Å) | | Cite: | Design of Potent and Highly Selective Inhibitors for Human beta-Secretase 2 (Memapsin 1), a Target for Type 2 Diabetes.
Chem Sci, 7, 2016
|
|
5XES
 
 | | TK9 NMR structure in SDS micelle | | Descriptor: | THR-VAL-TYR-VAL-TYR-SER-ARG-VAL-LYS | | Authors: | Ghosh, A, Bhunia, A. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights of a self-assembling 9-residue peptide from the C-terminal tail of the SARS corona virus E-protein in DPC and SDS micelles: A combined high and low resolution spectroscopic study.
Biochim Biophys Acta Biomembr, 1860, 2018
|
|
4ESO
 
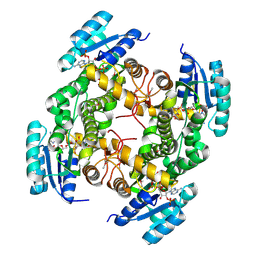 | | Crystal structure of a putative oxidoreductase protein from Sinorhizobium meliloti 1021 in complex with NADP | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | Authors: | Ghosh, A, Bhoshle, R, Toro, R, Gizzi, A, Hillerich, B, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-16 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Crystal structure of a putative oxidoreductase protein from Sinorhizobium meliloti 1021 in complex with NADP
To be Published
|
|
4GXH
 
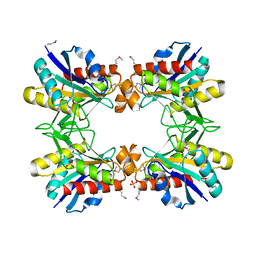 | |
4H16
 
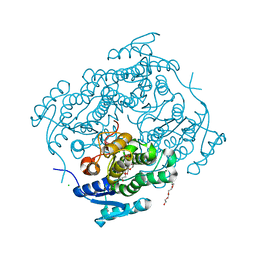 | |
4HKM
 
 | |
2N6M
 
 | |
4H15
 
 | |
4HPS
 
 | |
2MJX
 
 | | Solution NMR structure of a mismatch DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*AP*CP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*AP*TP*GP*CP*TP*AP*CP*GP*CP*G)-3') | | Authors: | Ghosh, A, Kumar, K.R, Bhunia, A, Chatterjee, S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Double GC:GC mismatch in dsDNA enhances local dynamics retaining the DNA footprint: a high-resolution NMR study
Chemmedchem, 9, 2014
|
|
9B2H
 
 | | HIV-1 wild type protease with GRL-072-17A, a substituted tetrahydrofuran derivative based on Darunavir as P2 group | | Descriptor: | 2,5:6,9-dianhydro-1,3,7,8-tetradeoxy-4-O-({(2S,3R)-3-hydroxy-4-[(4-methoxybenzene-1-sulfonyl)(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamoyl)-L-gluco-nonitol, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2024-03-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Design of substituted tetrahydrofuran derivatives for HIV-1 protease inhibitors: synthesis, biological evaluation, and X-ray structural studies.
Org.Biomol.Chem., 22, 2024
|
|
6UJ0
 
 | | Unbound BACE2 mutant structure | | Descriptor: | Beta-secretase 2, unidentified polypeptide | | Authors: | Yen, Y.C, Ghosh, A.K, Mesecar, A.D. | | Deposit date: | 2019-10-01 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Structure-Based Discovery Platform for BACE2 and the Development of Selective BACE Inhibitors.
Acs Chem Neurosci, 12, 2021
|
|
3BT4
 
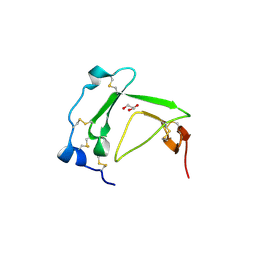 | | Crystal Structure Analysis of AmFPI-1, fungal protease inhibitor from Antheraea mylitta | | Descriptor: | Fungal protease inhibitor-1, GLYCEROL | | Authors: | Roy, S, Aravind, P, Madhurantakam, C, Ghosh, A.K, Sankarananarayanan, R, Das, A.K. | | Deposit date: | 2007-12-27 | | Release date: | 2008-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a fungal protease inhibitor from Antheraea mylitta
J.Struct.Biol., 166, 2009
|
|
7RC0
 
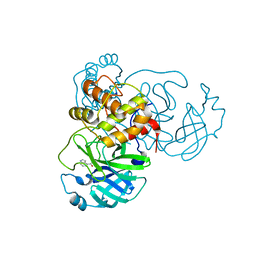 | | X-ray Structure of SARS-CoV-2 main protease covalently modified by compound GRL-091-20 | | Descriptor: | 3C-like proteinase, 5-chloro-4-methylpyridin-3-yl 1H-indole-4-carboxylate, SODIUM ION | | Authors: | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
7RBZ
 
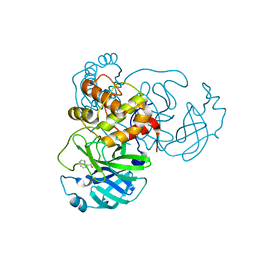 | | X-ray Structure of SARS-CoV-2 main protease covalently modified by compound GRL-017-20 | | Descriptor: | 3C-like proteinase, 5-chloropyridin-3-yl 2,3-dihydro-1H-indole-4-carboxylate | | Authors: | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
7RBW
 
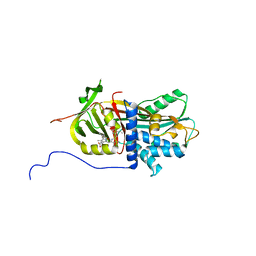 | | Structure of Biliverdin-binding Serpin of Boana punctata (polka-dot tree frog) | | Descriptor: | BILIVERDINE IX ALPHA, Biliverdin bindin serpin | | Authors: | Fedorov, E, Manoilov, K.Y, Verkhusha, V, Almo, S.C, Ghosh, A. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Functional Characterization of a Biliverdin-Binding Near-Infrared Fluorescent Protein From the Serpin Superfamily.
J.Mol.Biol., 434, 2021
|
|
7RC1
 
 | | X-ray Structure of SARS-CoV main protease covalently modified by compound GRL-0686 | | Descriptor: | 3C-like proteinase, 5-chloropyridin-3-yl 1-(3-nitrobenzene-1-sulfonyl)-1H-indole-5-carboxylate, DIMETHYL SULFOXIDE | | Authors: | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-07 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
8F0F
 
 | | HIV-1 wild type protease with GRL-110-19A, a chloroacetamide derivative based on Darunavir as P2' group | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[4-(2-chloroacetamido)benzene-1-sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2022-11-02 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Evaluation of darunavir-derived HIV-1 protease inhibitors incorporating P2' amide-derivatives: Synthesis, biological evaluation and structural studies.
Bioorg.Med.Chem.Lett., 83, 2023
|
|
8SWP
 
 | | Structure of K. lactis PNP bound to hypoxanthine | | Descriptor: | ACETATE ION, HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Fedorov, E, Ghosh, A. | | Deposit date: | 2023-05-19 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphate Binding in PNP Alters Transition-State Analogue Affinity and Subunit Cooperativity.
Biochemistry, 62, 2023
|
|
