1TPZ
 
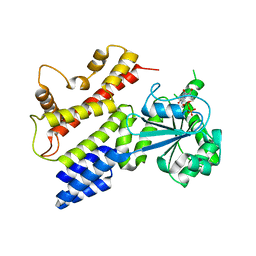 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | 登録日 | 2004-06-16 | | 公開日 | 2004-09-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
1TQ4
 
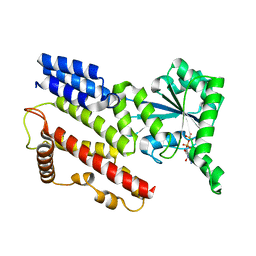 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, interferon-inducible GTPase | | 著者 | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | 登録日 | 2004-06-16 | | 公開日 | 2004-09-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
1TQ2
 
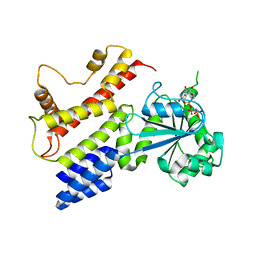 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | 著者 | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | 登録日 | 2004-06-16 | | 公開日 | 2004-09-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
1TQ6
 
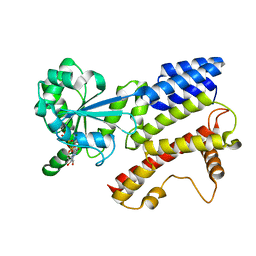 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, interferon-inducible GTPase | | 著者 | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | 登録日 | 2004-06-16 | | 公開日 | 2004-09-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
5DQC
 
 | | Co-crystal of BACE1 with compound 0211 | | 分子名称: | Beta-secretase 1, N-[(2S,3R)-3-hydroxy-4-({(2S,3S)-3-hydroxy-1-[(2-methylpropyl)amino]-1-oxobutan-2-yl}amino)-1-phenylbutan-2-yl]-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | 著者 | Ghosh, A.K, Bhavanam, S.R, Yen, T.-C, Cardenas, E.L, Rao, K.V, Downs, D, Huang, X, Tang, J, Mescar, A.D. | | 登録日 | 2015-09-14 | | 公開日 | 2016-02-17 | | 最終更新日 | 2016-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4651 Å) | | 主引用文献 | Design of Potent and Highly Selective Inhibitors for Human beta-Secretase 2 (Memapsin 1), a Target for Type 2 Diabetes.
Chem Sci, 7, 2016
|
|
4GID
 
 | | Structure of beta-secretase complexed with inhibitor | | 分子名称: | Beta-secretase 1, L-PROLINAMIDE, N-[(2S)-1-({(2S,3R)-3-hydroxy-1-[(2-methylpropyl)amino]-1-oxobutan-2-yl}amino)-3-phenylpropan-2-yl]-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | 著者 | Ghosh, A, Tang, J, Venkateswara, R.K, Yadav, N, Anderson, D, Gavande, N, Huang, X, Terzyan, S. | | 登録日 | 2012-08-08 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-based design of highly selective beta-secretase inhibitors: synthesis, biological evaluation, and protein-ligand X-ray crystal structure.
J.Med.Chem., 55, 2012
|
|
4HPS
 
 | |
3RTX
 
 | |
4H15
 
 | |
4HKM
 
 | |
4GXH
 
 | |
4H16
 
 | |
1XS7
 
 | | Crystal Structure of a cycloamide-urethane-derived novel inhibitor bound to human brain memapsin 2 (beta-secretase). | | 分子名称: | Beta-secretase 1, N-[(4S,5S,7R)-8-({(S)-1-[(BENZYLAMINO)OXOMETHYL]-2-METHYLPROPYL}AMINO)-5-HYDROXY-2,7-DIMETHYL-8-OXO-OCT-4-YL]-(4S,7S)-4 -ISOPROPYL-2,5,9-TRIOXO-1-OXA-3,6,10-TRIAZACYCLOHEXADECANE-7-CARBOXAMIDE | | 著者 | Ghosh, A, Devasamudram, T, Hong, L, DeZutter, C, Xu, X, Weerasena, V, Koelsch, G, Bilcer, G, Tang, J. | | 登録日 | 2004-10-18 | | 公開日 | 2004-12-21 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure-based design of cycloamide-urethane-derived novel inhibitors of human brain memapsin 2 (beta-secretase).
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2MJX
 
 | | Solution NMR structure of a mismatch DNA | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*TP*AP*CP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*AP*TP*GP*CP*TP*AP*CP*GP*CP*G)-3') | | 著者 | Ghosh, A, Kumar, K.R, Bhunia, A, Chatterjee, S. | | 登録日 | 2014-01-21 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Double GC:GC mismatch in dsDNA enhances local dynamics retaining the DNA footprint: a high-resolution NMR study
Chemmedchem, 9, 2014
|
|
2JR6
 
 | | Solution structure of UPF0434 protein NMA0874. Northeast Structural Genomics Target MR32 | | 分子名称: | UPF0434 protein NMA0874 | | 著者 | Ghosh, A, Singarapu, K.K, Wu, Y, Liu, G, Sukumaran, D, Chen, C.X, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-06-20 | | 公開日 | 2007-07-24 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of UPF0434 protein NMA0874.
To be Published
|
|
2N6M
 
 | |
3D20
 
 | | Crystal structure of HIV-1 mutant I54V and inhibitor DARUNAVIA | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, HIV-1 Protease, ... | | 著者 | Liu, F, Kovalesky, A.Y, Tie, Y, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | 登録日 | 2008-05-07 | | 公開日 | 2008-05-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
4ZFQ
 
 | | Structure of M. tuberculosis (3,3) L,D-Transpeptidase, LdtMt5. (Meropenen-adduct form) | | 分子名称: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, DI(HYDROXYETHYL)ETHER, L,D-transpeptidase 5 | | 著者 | Basta, L, Ghosh, A, Lamichhane, G, Bianchet, M.A. | | 登録日 | 2015-04-21 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.799 Å) | | 主引用文献 | Loss of a Functionally and Structurally Distinct ld-Transpeptidase, LdtMt5, Compromises Cell Wall Integrity in Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
5KAO
 
 | | Crystal structure of wild type HIV-1 protease in complex with GRL-10413 | | 分子名称: | [(3~{a}~{S},4~{R},6~{a}~{R})-2,3,3~{a},4,5,6~{a}-hexahydrofuro[2,3-b]furan-4-yl] ~{N}-[(2~{S},3~{R})-1-(3-chloranyl-4-methoxy-phenyl)-4-[(4-methoxyphenyl)sulfonyl-(2-methylpropyl)amino]-3-oxidanyl-butan-2-yl]carbamate, protease | | 著者 | Yedidi, R.S, Delino, N.S, Das, D, Kaufman, J.D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2016-06-01 | | 公開日 | 2016-08-31 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A Modified P1 Moiety Enhances In Vitro Antiviral Activity against Various Multidrug-Resistant HIV-1 Variants and In Vitro Central Nervous System Penetration Properties of a Novel Nonpeptidic Protease Inhibitor, GRL-10413.
Antimicrob.Agents Chemother., 60, 2016
|
|
1M4H
 
 | | Crystal Structure of Beta-secretase complexed with Inhibitor OM00-3 | | 分子名称: | Inhibitor OM00-3, beta-Secretase | | 著者 | Hong, L, Turner, R.T, Koelsch, G, Ghosh, A.K, Tang, J. | | 登録日 | 2002-07-02 | | 公開日 | 2002-08-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of Memapsin 2 (beta-Secretase) in Complex with Inhibitor OM00-3
Biochemistry, 41, 2002
|
|
7RXV
 
 | | Human Methionine Adenosyltransferase 2A bound to Methylthioadenosine, Malonate (MLA) and MgF3 | | 分子名称: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALANINE, GLYCEROL, ... | | 著者 | Fedorov, E, Niland, C.N, Schramm, V.L, Ghosh, A. | | 登録日 | 2021-08-23 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Mechanism of Triphosphate Hydrolysis by Human MAT2A at 1.07 angstrom Resolution.
J.Am.Chem.Soc., 143, 2021
|
|
7RXW
 
 | | Human Methionine Adenosyltransferase 2A bound to Methylthioadenosine and inhibitor imido-diphosphate (PNP) | | 分子名称: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALANINE, ... | | 著者 | Fedorov, E, Niland, C.N, Schramm, V.L, Ghosh, A. | | 登録日 | 2021-08-23 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Mechanism of Triphosphate Hydrolysis by Human MAT2A at 1.07 angstrom Resolution.
J.Am.Chem.Soc., 143, 2021
|
|
7RXX
 
 | | Human Methionine Adenosyltransferase 2A bound to Methylthioadenosine and two sulfate in the active site | | 分子名称: | 1,2-ETHANEDIOL, 3,6,9,12,15-PENTAOXAHEPTADECANE, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | 著者 | Fedorov, E, Niland, C.N, Schramm, V.L, Ghosh, A. | | 登録日 | 2021-08-23 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Mechanism of Triphosphate Hydrolysis by Human MAT2A at 1.07 angstrom Resolution.
J.Am.Chem.Soc., 143, 2021
|
|
7RJM
 
 | | Crystal structure of human Bromodomain containing protein 3 (BRD3) in complex with ILF3 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, Interleukin enhancer-binding factor 3 | | 著者 | Fedorov, E, Islam, K, Ghosh, A. | | 登録日 | 2021-07-21 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
Biorxiv, 2021
|
|
7RJL
 
 | | Crystal structure of human Bromodomain containing protein 3 (BRD3) in complex with SHMT | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, GLYCEROL, ... | | 著者 | Fedorov, E, Islam, K, Ghosh, A. | | 登録日 | 2021-07-21 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
Biorxiv, 2021
|
|
