2AV6
 
 | |
2AW3
 
 | |
5CX7
 
 | | Crystal Structure of PduOC:Heme Complex | | Descriptor: | ATP:cob(I)alamin adenosyltransferase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Geremia, S, Hickey, N, Ortiz de Orue Lucana, D. | | Deposit date: | 2015-07-28 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Crystal Structure of the C-Terminal Domain of the Salmonella enterica PduO Protein: An Old Fold with a New Heme-Binding Mode.
Front Microbiol, 7, 2016
|
|
6SUV
 
 | |
1EC5
 
 | | CRYSTAL STRUCTURE OF FOUR-HELIX BUNDLE MODEL | | Descriptor: | PROTEIN (FOUR-HELIX BUNDLE MODEL), ZINC ION | | Authors: | Geremia, S. | | Deposit date: | 2000-01-25 | | Release date: | 2000-07-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inaugural article: retrostructural analysis of metalloproteins: application to the design of a minimal model for diiron proteins.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FJ0
 
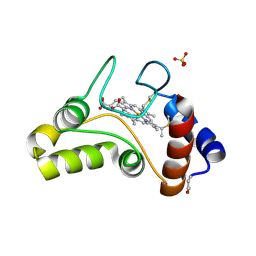 | |
2ASV
 
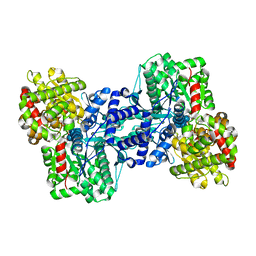 | |
2AZD
 
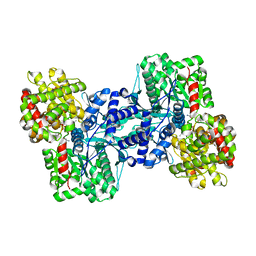 | |
3TQ2
 
 | |
1L5V
 
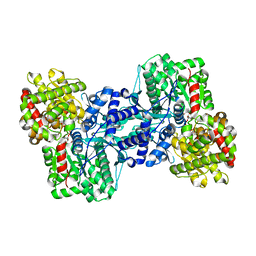 | | Crystal Structure of the Maltodextrin Phosphorylase complexed with Glucose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MALTODEXTRIN PHOSPHORYLASE, ... | | Authors: | Geremia, S, Campagnolo, M, Schinzel, R, Johnson, L.N. | | Deposit date: | 2002-03-08 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic catalysis in crystals of Escherichia coli maltodextrin phosphorylase
J.Mol.Biol., 322, 2002
|
|
1L6I
 
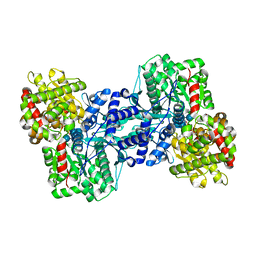 | | Crystal Structure of the Maltodextrin Phosphorylase complexed with the products of the enzymatic reaction between glucose-1-phosphate and maltopentaose | | Descriptor: | MALTODEXTRIN PHOSPHORYLASE, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Geremia, S, Campagnolo, M, Schinzel, R, Johnson, L.N. | | Deposit date: | 2002-03-11 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic catalysis in crystals of Escherichia coli maltodextrin phosphorylase
J.Mol.Biol., 322, 2002
|
|
1L5W
 
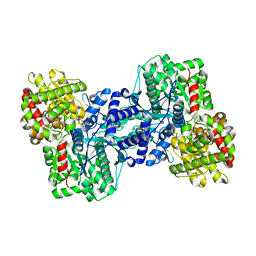 | | Crystal Structure of the Maltodextrin Phosphorylase Complexed with the Products of the Enzymatic Reaction between Glucose-1-phosphate and Maltotetraose | | Descriptor: | MALTODEXTRIN PHOSPHORYLASE, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Geremia, S, Campagnolo, M, Schinzel, R, Johnson, L.N. | | Deposit date: | 2002-03-08 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzymatic catalysis in crystals of Escherichia coli maltodextrin phosphorylase
J.Mol.Biol., 322, 2002
|
|
3TOH
 
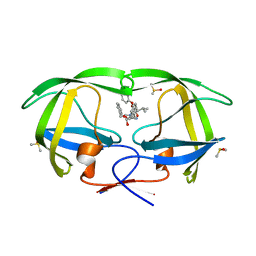 | | HIV-1 Protease - Epoxydic Inhibitor Complex (pH 9 - Orthorombic Crystal form P212121) | | Descriptor: | (S)-N-((2S,3S,4R,5R)-4-amino-3,5-dihydroxy-1,6-diphenylhexan-2-yl)-3-methyl-2-(2-phenoxyacetamido)butanamide, DIMETHYL SULFOXIDE, Gag-Pol polyprotein | | Authors: | Geremia, S, Olajuyigbe, F.M, Demitri, N. | | Deposit date: | 2011-09-05 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.116 Å) | | Cite: | Developing HIV-1 Protease Inhibitors through Stereospecific Reactions in Protein Crystals.
Molecules, 21, 2016
|
|
3TOG
 
 | | HIV-1 Protease - Epoxydic Inhibitor Complex (pH 9 - Monoclinic Crystal form P21) | | Descriptor: | (S)-N-((2S,3S,4R,5R)-4-amino-3,5-dihydroxy-1,6-diphenylhexan-2-yl)-3-methyl-2-(2-phenoxyacetamido)butanamide, DIMETHYL SULFOXIDE, Gag-Pol polyprotein | | Authors: | Geremia, S, Olajuyigbe, F.M, Demitri, N. | | Deposit date: | 2011-09-05 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Developing HIV-1 Protease Inhibitors through Stereospecific Reactions in Protein Crystals.
Molecules, 21, 2016
|
|
3TOF
 
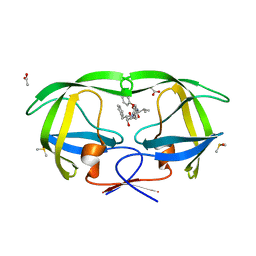 | | HIV-1 Protease - Epoxydic Inhibitor Complex (pH 6 - Orthorombic Crystal form P212121) | | Descriptor: | (S)-N-((1R,2S)-1-((2R,3R)-3-benzyloxiran-2-yl)-1-hydroxy-3-phenylpropan-2-yl)-3-methyl-2-(2-phenoxyacetamido)butanamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Geremia, S, Olajuyigbe, F.M, Ajele, J.O, Demitri, N, Randaccio, L, Wuerges, J, Benedetti, L, Campaner, P, Berti, F. | | Deposit date: | 2011-09-05 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Developing HIV-1 Protease Inhibitors through Stereospecific Reactions in Protein Crystals.
Molecules, 21, 2016
|
|
3NDU
 
 | | HIV-1 Protease Saquinavir:Ritonavir 1:5 complex structure | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Geremia, S, Olajuyigbe, F.M, Demitri, N. | | Deposit date: | 2010-06-08 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Investigation of 2-Fold Disorder of Inhibitors and Relative Potency by Crystallizations of HIV-1 Protease in Ritonavir and Saquinavir Mixtures
Cryst.Growth Des., 11, 2011
|
|
3NDW
 
 | | HIV-1 Protease Saquinavir:Ritonavir 1:15 complex structure | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Protease, ... | | Authors: | Geremia, S, Olajuyigbe, F.M, Demitri, N. | | Deposit date: | 2010-06-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Investigation of 2-Fold Disorder of Inhibitors and Relative Potency by Crystallizations of HIV-1 Protease in Ritonavir and Saquinavir Mixtures
Cryst.Growth Des., 11, 2011
|
|
3NDT
 
 | | HIV-1 Protease Saquinavir:Ritonavir 1:1 complex structure | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Geremia, S, Olajuyigbe, F.M, Demitri, N. | | Deposit date: | 2010-06-08 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Investigation of 2-Fold Disorder of Inhibitors and Relative Potency by Crystallizations of HIV-1 Protease in Ritonavir and Saquinavir Mixtures
Cryst.Growth Des., 11, 2011
|
|
3NDX
 
 | | HIV-1 Protease Saquinavir:Ritonavir 1:50 complex structure | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Geremia, S, Olajuyigbe, F.M, Demitri, N. | | Deposit date: | 2010-06-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Investigation of 2-Fold Disorder of Inhibitors and Relative Potency by Crystallizations of HIV-1 Protease in Ritonavir and Saquinavir Mixtures
Cryst.Growth Des., 11, 2011
|
|
3K4V
 
 | | New crystal form of HIV-1 Protease/Saquinavir structure reveals carbamylation of N-terminal proline | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Olajuyigbe, F.M, Demitri, N, Ajele, J.O, Maurizio, E, Randaccio, L, Geremia, S. | | Deposit date: | 2009-10-06 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Carbamylation of N-terminal proline.
ACS Med Chem Lett, 1, 2010
|
|
1QM5
 
 | | Phosphorylase recognition and phosphorylysis of its oligosaccharide substrate: answers to a long outstanding question | | Descriptor: | 4-thio-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, MALTODEXTRIN PHOSPHORYLASE, PHOSPHATE ION, ... | | Authors: | Watson, K.A, McCleverty, C, Geremia, S, Cottaz, S, Driguez, H, Johnson, L.N. | | Deposit date: | 1999-09-20 | | Release date: | 2000-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylase Recognition and Phosphorolysis of its Oligosaccharide Substrate: Answers to a Long Outstanding Question
Embo J., 18, 1999
|
|
2A1E
 
 | | High resolution structure of HIV-1 PR with TS-126 | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Demitri, N, Geremia, S, Randaccio, L, Wuerges, J, Benedetti, F, Berti, F, Dinon, F, Campaner, P, Tell, G. | | Deposit date: | 2005-06-20 | | Release date: | 2006-02-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A potent HIV protease inhibitor identified in an epimeric mixture by high-resolution protein crystallography.
Chemmedchem, 1, 2006
|
|
1OVU
 
 | |
1OVV
 
 | |
1OVR
 
 | | CRYSTAL STRUCTURE OF FOUR-HELIX BUNDLE MODEL di-Mn(II)-DF1-L13 | | Descriptor: | MANGANESE (II) ION, four-helix bundle model di-Mn(II)-DF1-L13 | | Authors: | Di Costanzo, L, Geremia, S. | | Deposit date: | 2003-03-27 | | Release date: | 2004-05-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Response of a designed metalloprotein to changes in metal ion coordination, exogenous ligands, and active site volume determined by X-ray crystallography.
J.Am.Chem.Soc., 127, 2005
|
|
