5K35
 
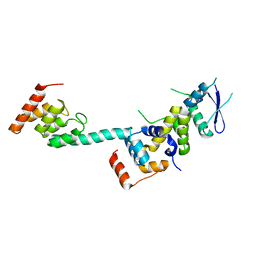 | | Structure of the Legionella effector, AnkB, in complex with human Skp1 | | Descriptor: | Ankyrin-repeat protein B, S-phase kinase-associated protein 1 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-05-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Mimicry by a Bacterial F Box Effector Hijacks the Host Ubiquitin-Proteasome System.
Structure, 25, 2017
|
|
5K34
 
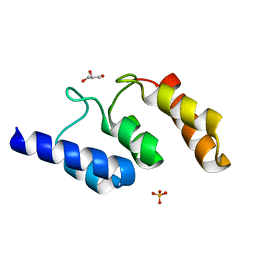 | | Structure of the ankyrin domain of AnkB from Legionella Pneumophila | | Descriptor: | Ankyrin-repeat protein B, GLYCEROL, SULFATE ION | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-05-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Mimicry by a Bacterial F Box Effector Hijacks the Host Ubiquitin-Proteasome System.
Structure, 25, 2017
|
|
5KDG
 
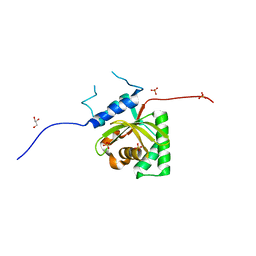 | | Crystal Structure of Salmonella Typhimurium Effector GtgE | | Descriptor: | GLYCEROL, Gifsy-2 prophage protein, SULFATE ION | | Authors: | Kozlov, G, Xu, C, Wong, K, Gehring, K, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-06-08 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the Salmonella Typhimurium Effector GtgE.
PLoS ONE, 11, 2016
|
|
4JU5
 
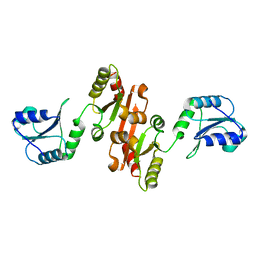 | |
4JRD
 
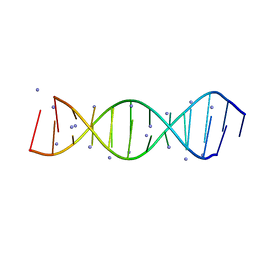 | | Crystal structure of the parallel double-stranded helix of poly(A) RNA | | Descriptor: | AMMONIUM ION, RNA (5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Safaee, N, Noronha, A.M, Kozlov, G, Rodionov, D, Wilds, C.J, Sheldrick, G.M, Gehring, K. | | Deposit date: | 2013-03-21 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of the parallel duplex of poly(A) RNA: evaluation of a 50 year-old prediction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
5KES
 
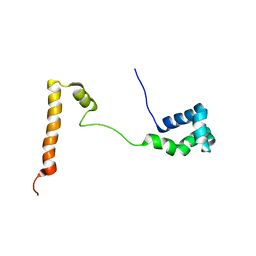 | | Solution structure of the yeast Ddi1 HDD domain | | Descriptor: | DNA damage-inducible protein 1 | | Authors: | Trempe, J.-F, Ratcliffe, C, Veverka, V, Saskova, K, Gehring, K. | | Deposit date: | 2016-06-10 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the yeast DNA damage-inducible protein Ddi1 reveal domain architecture of this eukaryotic protein family.
Sci Rep, 6, 2016
|
|
3IQL
 
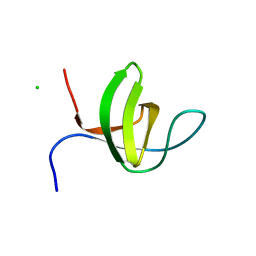 | | Crystal structure of the rat endophilin-A1 SH3 domain | | Descriptor: | CHLORIDE ION, Endophilin-A1 | | Authors: | Trempe, J.F, Kozlov, G, Camacho, E.M, Gehring, K. | | Deposit date: | 2009-08-20 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | SH3 domains from a subset of BAR proteins define a Ubl-binding domain and implicate parkin in synaptic ubiquitination.
Mol.Cell, 36, 2009
|
|
1P9K
 
 | | THE SOLUTION STRUCTURE OF YBCJ FROM E. COLI REVEALS A RECENTLY DISCOVERED ALFAL MOTIF INVOLVED IN RNA-BINDING | | Descriptor: | orf, hypothetical protein | | Authors: | Volpon, L, Lievre, C, Osborne, M.J, Gandhi, S, Iannuzzi, P, Larocque, R, Matte, A, Cygler, M, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-05-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of YbcJ from Escherichia coli reveals a recently discovered alphaL motif involved in RNA binding.
J.Bacteriol., 185, 2003
|
|
1NEE
 
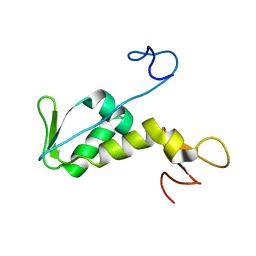 | | Structure of archaeal translation factor aIF2beta from Methanobacterium thermoautrophicum | | Descriptor: | Probable translation initiation factor 2 beta subunit, ZINC ION | | Authors: | Gutierrez, P, Trempe, J.F, Siddiqui, N, Arrowsmith, C, Gehring, K. | | Deposit date: | 2002-12-11 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the archaeal translation initiation factor aIF2beta from Methanobacterium thermoautotrophicum: Implications for translation initiation.
Protein Sci., 13, 2004
|
|
1NMR
 
 | | Solution Structure of C-terminal Domain from Trypanosoma cruzi Poly(A)-Binding Protein | | Descriptor: | poly(A)-binding protein | | Authors: | Siddiqui, N, Kozlov, G, D'Orso, I, Trempe, J.F, Frasch, A.C.C, Gehring, K. | | Deposit date: | 2003-01-10 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain from poly(A)-binding protein in Trypanosoma cruzi: A vegetal PABC domain
Protein Sci., 12, 2003
|
|
1ME1
 
 | | Chimeric hairpin with 2',5'-linked RNA loop and RNA stem | | Descriptor: | 5'-R(*GP*GP*AP*CP*(U25)P*(U25)P*(C25)P*(5GP)P*GP*UP*CP*C)-3' | | Authors: | Denisov, A.Y, Hannoush, R.N, Gehring, K, Damha, M.J. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Novel RNA Motif Based on the Structure of Unusually Stable 2',5'-Linked r(UUCG) Loops
J.AM.CHEM.SOC., 125, 2003
|
|
1MK3
 
 | | SOLUTION STRUCTURE OF HUMAN BCL-W PROTEIN | | Descriptor: | Apoptosis regulator Bcl-W | | Authors: | Denisov, A.Y, Madiraju, M.S, Chen, G, Khadir, A, Beauparlant, P, Attardo, G, Shore, G.C, Gehring, K. | | Deposit date: | 2002-08-28 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human BCL-w: modulation of ligand binding by the C-terminal helix
J.BIOL.CHEM., 278, 2003
|
|
1ME0
 
 | | Chimeric hairpin with 2',5'-linked RNA loop and DNA stem | | Descriptor: | DNA/RNA (5'-D(*GP*GP*AP*C)-R(P*(U25)P*(U25)P*(C25)P*(5GP))-D(P*GP*TP*CP*C)-3') | | Authors: | Denisov, A.Y, Hannoush, R.N, Gehring, K, Damha, M.J. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Novel RNA Motif Based on the Structure of Unusually Stable 2',5'-Linked r(UUCG) Loops
J.AM.CHEM.SOC., 125, 2003
|
|
4EEW
 
 | |
4G3O
 
 | |
1JGN
 
 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip2 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein 2 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-26 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
1JH4
 
 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip1 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein-1 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-27 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
1D5G
 
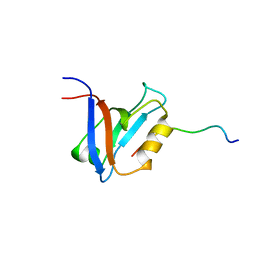 | |
5BTZ
 
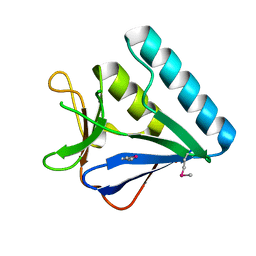 | | Structure of the middle domain of lpg1496 from Legionella pneumophila in P212121 space group | | Descriptor: | lpg1496 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5BTX
 
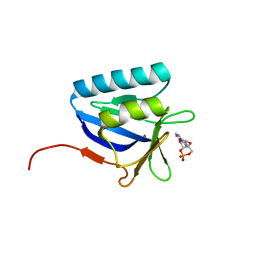 | | Structure of the N-terminal domain of lpg1496 from Legionella pneumophila in complex with nucleotide | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, lpg1496 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
1YHD
 
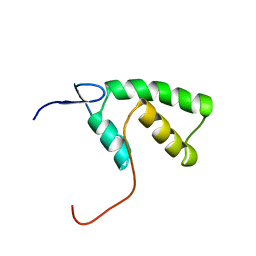 | | The solution structure of YGGX from Escherichia Coli | | Descriptor: | UPF0269 protein yggX | | Authors: | Osborne, M.J, Siddiqui, N, Landgraf, D, Pomposiello, P.J, Gehring, K. | | Deposit date: | 2005-01-07 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the oxidative stress-related protein YggX from Escherichia coli.
Protein Sci., 14, 2005
|
|
5BU0
 
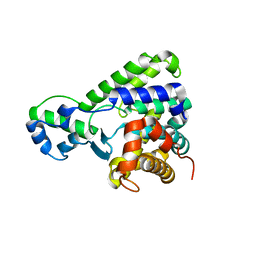 | | Structure of the C-terminal domain of lpg1496 from Legionella pneumophila | | Descriptor: | lpg1496 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5BTW
 
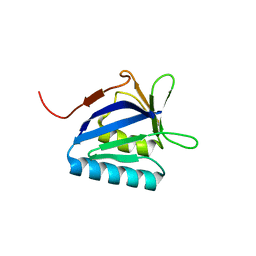 | | Structure of the N-terminal domain of lpg1496 from Legionella pneumophila | | Descriptor: | Uncharacterized protein | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5BU2
 
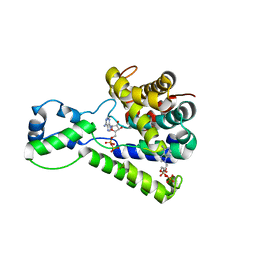 | | Structure of the C-terminal domain of lpg1496 from Legionella pneumophila in complex with nucleotide | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, alpha-D-ribofuranose, ... | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
1Y00
 
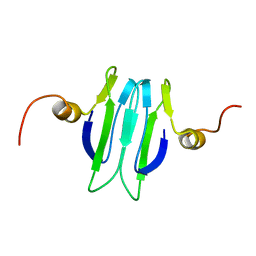 | | Solution structure of the Carbon Storage Regulator protein CsrA | | Descriptor: | Carbon storage regulator | | Authors: | Gutierrez, P, Li, Y, Osborne, M.J, Liu, Q, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-11-13 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the carbon storage regulator protein CsrA from Escherichia coli.
J.Bacteriol., 187, 2005
|
|
