3NPH
 
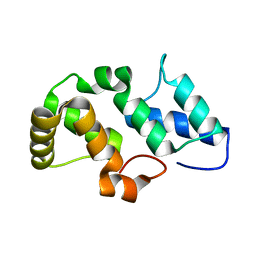 | | Crystal structure of the pfam00427 domain from Synechocystis sp. PCC 6803 | | Descriptor: | Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, rod 2 | | Authors: | Gao, X, Chen, L, Wu, J.-W, Zhang, Y.-Z. | | Deposit date: | 2010-06-28 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Crystal structure of the N-terminal domain of linker L(R) and the assembly of cyanobacterial phycobilisome rods
Mol.Microbiol., 82, 2011
|
|
8X77
 
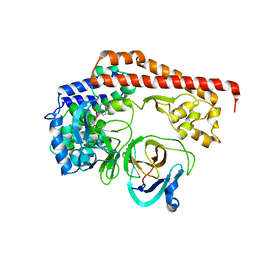 | | Enterovirus proteinase with host factor | | Descriptor: | 2A protein, Actin-histidine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2023-11-23 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | The EV71 2A protease occupies the central cleft of SETD3 and disrupts SETD3-actin interaction.
Nat Commun, 15, 2024
|
|
5ZT1
 
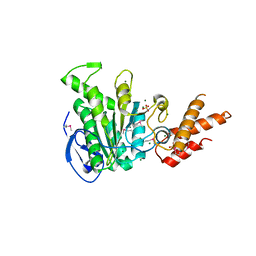 | | Structure of the bacterial pathogens ATPase with substrate ATP gamma S | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Probable ATP synthase SpaL/MxiB, ... | | Authors: | Gao, X.P, Mu, Z.X, Cui, S. | | Deposit date: | 2018-05-01 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.114 Å) | | Cite: | Structural Insight Into Conformational Changes Induced by ATP Binding in a Type III Secretion-Associated ATPase FromShigella flexneri
Front Microbiol, 9, 2018
|
|
7DHG
 
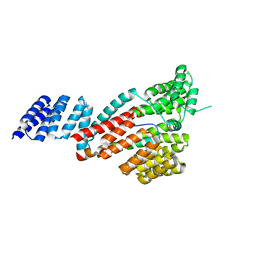 | | Crystal structure of SARS-CoV-2 Orf9b complex with human TOM70 | | Descriptor: | Mitochondrial import receptor subunit TOM70, ORF9b protein | | Authors: | Gao, X, Zhu, K, Qin, B, Olieric, V, Wang, M, Cui, S. | | Deposit date: | 2020-11-14 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 Orf9b in complex with human TOM70 suggests unusual virus-host interactions.
Nat Commun, 12, 2021
|
|
7F60
 
 | |
7F90
 
 | |
7CJD
 
 | |
8IT0
 
 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA dimer (conformation-2) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8ISZ
 
 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA monomer | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8ISY
 
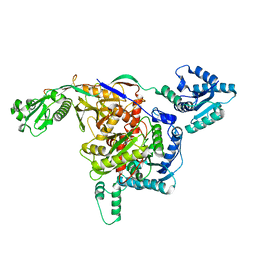 | | Cryo-EM structure of free-state Crt-SPARTA | | Descriptor: | Piwi domain-containing protein, TIR domain-containing protein | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8IT1
 
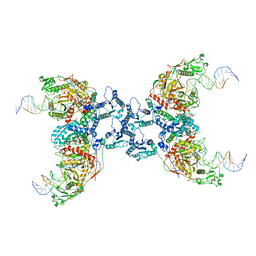 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA tetramer (NADase active form) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
7CMD
 
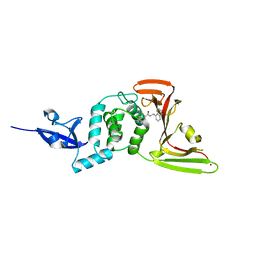 | | Crystal structure of the SARS-CoV-2 PLpro with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease.
Acta Pharm Sin B, 11, 2021
|
|
7CDW
 
 | |
6PWC
 
 | | A complex structure of arrestin-2 bound to neurotensin receptor 1 | | Descriptor: | Beta-arrestin-1, Fab30 heavy chain, Fab30 light chain, ... | | Authors: | Yin, W, Li, Z, Jin, M, Yin, Y.-L, de Waal, P.W, Pal, K, Gao, X, He, Y, Gao, J, Wang, X, Zhang, Y, Zhou, H, Melcher, K, Jiang, Y, Cong, Y, Zhou, X.E, Yu, X, Xu, H.E. | | Deposit date: | 2019-07-22 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | A complex structure of arrestin-2 bound to a G protein-coupled receptor.
Cell Res., 29, 2019
|
|
1R10
 
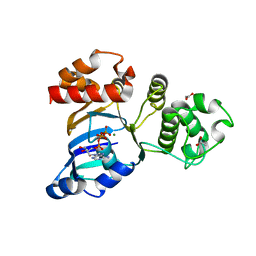 | | Cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domain one (NBD1) with ATP, I4122 space group | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Lewis, H.A, Buchanan, S.G, Burley, S.K, Conners, K, Dickey, M, Dorwart, M, Fowler, R, Gao, X, Guggino, W.B, Hendrickson, W.A. | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator.
Embo J., 23, 2004
|
|
1R0Z
 
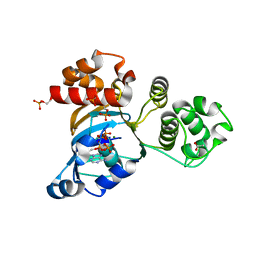 | | Phosphorylated Cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domain one (NBD1) with ATP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Lewis, H.A, Buchanan, S.G, Burley, S.K, Conners, K, Dickey, M, Dorwart, M, Fowler, R, Gao, X, Guggino, W.B, Hendrickson, W.A. | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator.
Embo J., 23, 2004
|
|
1R0Y
 
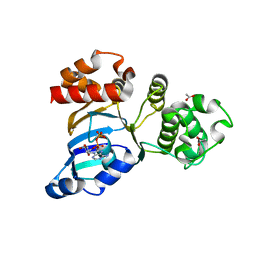 | | Cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domain one (NBD1) with ADP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator | | Authors: | Lewis, H.A, Buchanan, S.G, Burley, S.K, Conners, K, Dickey, M, Dorwart, M, Fowler, R, Gao, X, Guggino, W.B, Hendrickson, W.A. | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator.
Embo J., 23, 2004
|
|
1R0X
 
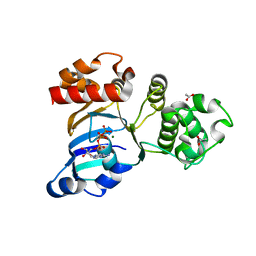 | | Cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domain one (NBD1) with ATP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Lewis, H.A, Buchanan, S.G, Burley, S.K, Conners, K, Dickey, M, Dorwart, M, Fowler, R, Gao, X, Guggino, W.B, Hendrickson, W.A. | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator.
Embo J., 23, 2004
|
|
1R0W
 
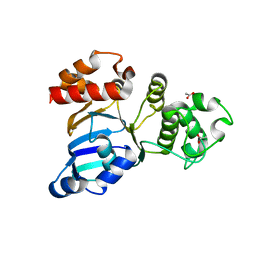 | | Cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domain one (NBD1) apo | | Descriptor: | ACETIC ACID, Cystic fibrosis transmembrane conductance regulator | | Authors: | Lewis, H.A, Buchanan, S.G, Burley, S.K, Conners, K, Dickey, M, Dorwart, M, Fowler, R, Gao, X, Guggino, W.B, Hendrickson, W.A. | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator.
Embo J., 23, 2004
|
|
6UH3
 
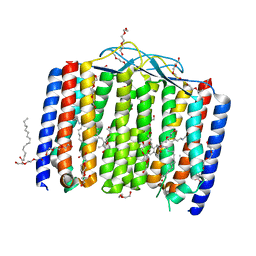 | | Crystal structure of bacterial heliorhodopsin 48C12 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Heliorhodopsin, PALMITIC ACID, ... | | Authors: | Lu, Y, Zhou, X.E, Gao, X, Xia, R, Xu, Z, Wang, N, Leng, Y, Melcher, K, Xu, H.E, He, Y. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of heliorhodopsin 48C12.
Cell Res., 30, 2020
|
|
8Y1R
 
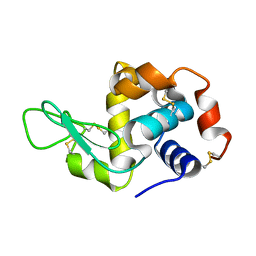 | | in situ room temperature Laue crystallography | | Descriptor: | Lysozyme C | | Authors: | Wang, Z.J, Wang, S.S, Pan, Q.Y, Yu, L, Su, Z.H, Yang, T.Y, Wang, Y.Z, Zhang, W.Z, Hao, Q, Gao, X.Y. | | Deposit date: | 2024-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single pulse data collection with an X-ray chopper at in situ room temperature Laue crystallography beamline BL03HB
Nucl Instrum Methods Phys Res A, 1069, 2024
|
|
7X1R
 
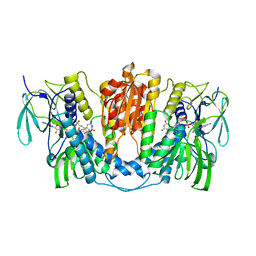 | | Cryo-EM structure of human thioredoxin reductase bound by Au | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, Thioredoxin reductase 1, ... | | Authors: | He, Z.S, Cao, P, Cao, S.H, He, B, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2022-02-24 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Au4 cluster inhibits human thioredoxin reductase activity via specifically binding of Au to Cys189
Nano Today, 47, 2022
|
|
7XBO
 
 | | Crystal Structure of 10-dml-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | (3R,4S,5S,7R,9E,11R,12R)-12-ETHYL-4-HYDROXY-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, Cytochrome P450 monooxygenase PikC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | New mechanistic insight of cytochrome P450 PikC gained from site-specific mutagenesis by non-coding amino acids
Nat Commun, 2023
|
|
7XBN
 
 | | Crystal Structure of YC-17-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, Cytochrome P450 monooxygenase PikC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New mechanistic insight of cytochrome P450 PikC gained from site-specific mutagenesis by non-coding amino acids
Nat Commun, 2023
|
|
7XBM
 
 | | Crystal Structure of cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | CACODYLATE ION, Cytochrome P450 monooxygenase PikC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238
Nat Commun, 2023
|
|
