6M4J
 
 | | SspA in complex with cysteine | | Descriptor: | CYSTEINE, PYRIDOXAL-5'-PHOSPHATE, SspA complex protein | | Authors: | Liu, L, Gao, H. | | Deposit date: | 2020-03-07 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of an l-Cysteine Desulfurase from an Ssp DNA Phosphorothioation System.
Mbio, 11, 2020
|
|
7YLL
 
 | | Crystal structure of TTEDbh | | Descriptor: | DNA polymerase IV, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Yan, X, Tian, L, Gao, H. | | Deposit date: | 2022-07-26 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6000092 Å) | | Cite: | Structure and function of extreme TLS DNA polymerase TTEDbh from Thermoanaerobacter tengcongensis.
Int.J.Biol.Macromol., 253, 2023
|
|
8XJ8
 
 | | The Cryo-EM structure of MPXV E5 C-terminal in complex with DNA | | Descriptor: | DNA (70-MER), MAGNESIUM ION, Monkeypox virus E5, ... | | Authors: | Zhang, W, Liu, Y, Gao, H, Gan, J. | | Deposit date: | 2023-12-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
8XJ6
 
 | | The Cryo-EM structure of MPXV E5 apo conformation | | Descriptor: | AMP PHOSPHORAMIDATE, Monkeypox virus E5, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhang, W, Liu, Y, Gao, H, Gan, J. | | Deposit date: | 2023-12-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
8XJ7
 
 | | The Cryo-EM structure of MPXV E5 in complex with DNA | | Descriptor: | DNA (70-MER), MAGNESIUM ION, Monkeypox virus E5, ... | | Authors: | Zhang, W, Liu, Y, Gao, H, Gan, J. | | Deposit date: | 2023-12-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
3TL8
 
 | |
4HPZ
 
 | |
8IZP
 
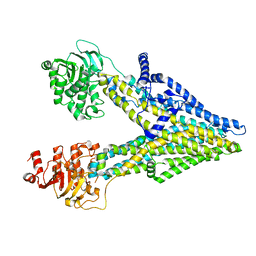 | | Multidrug resistance-associated protein 3 | | Descriptor: | ATP-binding cassette sub-family C member 3 | | Authors: | Yun, C.H, Gao, H.M. | | Deposit date: | 2023-04-07 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | structure of multidrug resistance-associated protein 3 at 3.31 Angstroms
To Be Published
|
|
8IZR
 
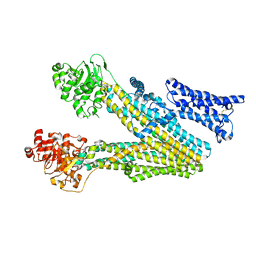 | | Multidrug resistance-associated protein 3 | | Descriptor: | ATP-binding cassette sub-family C member 2 | | Authors: | Yun, C.H, Gao, H.M. | | Deposit date: | 2023-04-07 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | structure of multidrug resistance-associated protein 2 at 3.62 Angstroms resolution
To Be Published
|
|
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2808516 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | Authors: | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7DR8
 
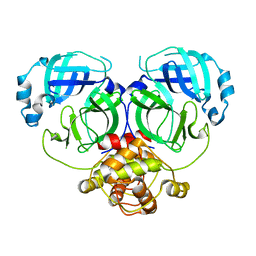 | | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121 | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-26 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.338149 Å) | | Cite: | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121
To Be Published
|
|
7XYF
 
 | |
7XYG
 
 | |
7XXE
 
 | |
