5YXA
 
 | | Crystal structure of the C-terminal fragment of NS1 protein from yellow fever virus | | Descriptor: | Non-structural protein 1 | | Authors: | Wang, H, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-12-04 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the C-terminal fragment of NS1 protein from yellow fever virus.
Sci China Life Sci, 60, 2017
|
|
5XL5
 
 | |
5XL3
 
 | | Complex structure of H4 hemagglutinin from avian influenza H4N6 virus with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-10 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation by Influenza A Virus Hemagglutinin H4
Cell Rep, 20, 2017
|
|
5XSQ
 
 | | Crystal Structure of the Marburg Virus Nucleoprotein Core Domain Chaperoned by a VP35 Peptide | | Descriptor: | Nucleoprotein, Peptide from Polymerase cofactor VP35 | | Authors: | Zhu, T, Song, H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-06-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Marburg Virus Nucleoprotein Core Domain Chaperoned by a VP35 Peptide Reveals a Conserved Drug Target for Filovirus
J. Virol., 91, 2017
|
|
5X5F
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 2 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5V
 
 | | Crystal structure of pseudorabies virus glycoprotein D | | Descriptor: | GD | | Authors: | Li, A, Lu, G, Qi, J, Wu, L, Tian, K, Luo, T, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2017-02-17 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of pseudorabies virus glycoprotein D
To Be Published
|
|
7XQT
 
 | | The structure of FLA-K*00701/KP-FECV-11 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, peptide from Spike glycoprotein | | Authors: | Qiao, P.W, Yue, C, Peng, W.Y, Liu, K.F, Huo, S.T, Zhang, D, Chai, Y, Qi, J.X, Sun, Z.Y, Gao, G.F, Liu, W.J, Wu, G.Z. | | Deposit date: | 2022-05-08 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the characteristics of feline major histocompatibility complex class I molecules cross-presenting coronavirus peptides
To Be Published
|
|
7XQS
 
 | | The structure of FLA-K*00701/KP-CoV-9 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, peptide from Spike glycoprotein | | Authors: | Qiao, P.W, Yue, C, Peng, W.Y, Liu, K.F, Huo, S.T, Zhang, D, Chai, Y, Qi, J.X, Sun, Z.Y, Gao, G.F, Liu, W.J, Wu, G.Z. | | Deposit date: | 2022-05-08 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Analysis of the characteristics of feline major histocompatibility complex class I molecules cross-presenting coronavirus peptides
To Be Published
|
|
7XQU
 
 | | The structure of FLA-E*00301/EM-FECV-10 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide from Nucleoprotein | | Authors: | Qiao, P.W, Yue, C, Peng, W.Y, Liu, K.F, Huo, S.T, Zhang, D, Chai, Y, Qi, J.X, Sun, Z.Y, Gao, G.F, Liu, W.J, Wu, G.Z. | | Deposit date: | 2022-05-08 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Analysis of the characteristics of feline major histocompatibility complex class I molecules cross-presenting coronavirus peptides
To Be Published
|
|
5XLC
 
 | |
6AEE
 
 | | Crystal structure of the four Ig-like domains of LILRB1 complexed with HLA-G | | Descriptor: | 9 Mer Peptide (RL9) From Histone H2A.x, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Wang, Q, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2018-08-04 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Structures of the four Ig-like domain LILRB2 and the four-domain LILRB1 and HLA-G1 complex.
Cell. Mol. Immunol., 2019
|
|
6A7W
 
 | | Structure of a catalytic domain of the colistin resistance enzyme | | Descriptor: | Putative integral membrane protein, ZINC ION | | Authors: | Wang, X.D, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2018-07-04 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.988 Å) | | Cite: | Structural and functional insights into MCR-2 mediated colistin resistance.
Sci China Life Sci, 61, 2018
|
|
6AED
 
 | |
5XGR
 
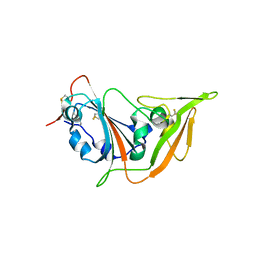 | | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Xue, H, Qi, J, Song, H, Qihui, W, Shi, Y, Gao, G.F. | | Deposit date: | 2017-04-16 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein
Virology, 507, 2017
|
|
5X4S
 
 | | Structure of the N-terminal domain (NTD)of SARS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5WZ3
 
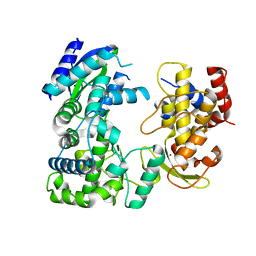 | | Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase(RdRP) | | Descriptor: | NS5 RdRp, ZINC ION | | Authors: | Duan, W, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | The crystal structure of Zika virus NS5 reveals conserved drug targets.
EMBO J., 36, 2017
|
|
5WXD
 
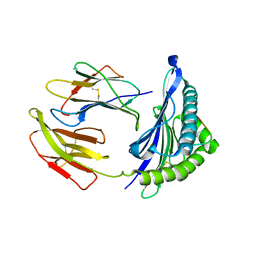 | | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1) | | Descriptor: | Beta-2-microglobulin, H7-25, HLA class I histocompatibility antigen, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-07 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Heterosubtypic Protections against Human-Infecting Avian Influenza Viruses Correlate to Biased Cross-T-Cell Responses.
Mbio, 9, 2018
|
|
5X4R
 
 | | Structure of the N-terminal domain (NTD) of MERS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X58
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5XEB
 
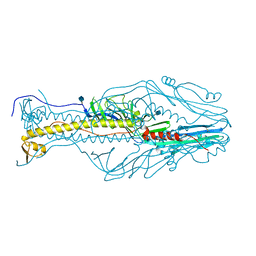 | | Structure of the envelope glycoprotein of Dhori virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein | | Authors: | Peng, R, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-04-03 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structures of human-infectingThogotovirusfusogens support a common ancestor with insect baculovirus
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5X5B
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 2 | | Descriptor: | Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5W
 
 | | Crystal structure of pseudorabies virus glycoprotein D | | Descriptor: | GD, Nectin-1 | | Authors: | Li, A, Lu, G, Qi, J, Wu, L, Tian, K, Luo, T, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2017-02-17 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of pseudorabies virus glycoprotein D
To Be Published
|
|
5XL7
 
 | |
5XL4
 
 | |
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
