1ZJK
 
 | | Crystal structure of the zymogen catalytic region of human MASP-2 | | Descriptor: | Mannan-binding lectin serine protease 2 | | Authors: | Gal, P, Harmat, V, Kocsis, A, Bian, T, Barna, L, Ambrus, G, Vegh, B, Balczer, J, Sim, R.B, Naray-Szabo, G, Zavodszky, P. | | Deposit date: | 2005-04-29 | | Release date: | 2005-07-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A True Autoactivating Enzyme: Structural insight into mannose-binding lectin-associated serine protease-2 activations
J.Biol.Chem., 280, 2005
|
|
3ZKE
 
 | | Structure of LC8 in complex with Nek9 peptide | | Descriptor: | DYNEIN LIGHT CHAIN 1, CYTOPLASMIC, NEK9 PROTEIN | | Authors: | Gallego, P, Velazquez-Campoy, A, Regue, L, Roig, J, Reverter, D. | | Deposit date: | 2013-01-22 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of the Regulation of the Dynll/Lc8 Binding to Nek9 by Phosphorylation
J.Biol.Chem., 288, 2013
|
|
3ZKF
 
 | | Structure of LC8 in complex with Nek9 phosphopeptide | | Descriptor: | DYNEIN LIGHT CHAIN 1, CYTOPLASMIC, NEK9 PROTEIN | | Authors: | Gallego, P, Velazquez-Campoy, A, Regue, L, Roig, J, Reverter, D. | | Deposit date: | 2013-01-22 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of the Regulation of the Dynll/Lc8 Binding to Nek9 by Phosphorylation
J.Biol.Chem., 288, 2013
|
|
7QCL
 
 | | Structure of the MUCIN-2 Cterminal domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mucin-2, ... | | Authors: | Gallego, P, Hansson, G.C. | | Deposit date: | 2021-11-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | The intestinal MUC2 mucin C-terminus is stabilized by an extra disulfide bond in comparison to von Willebrand factor and other gel-forming mucins.
Nat Commun, 14, 2023
|
|
7QCU
 
 | | Structure of the MUCIN-2 Cterminal domains partially deglycosylated. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mucin-2, ... | | Authors: | Gallego, P, Hansson, G.C. | | Deposit date: | 2021-11-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The intestinal MUC2 mucin C-terminus is stabilized by an extra disulfide bond in comparison to von Willebrand factor and other gel-forming mucins.
Nat Commun, 14, 2023
|
|
7QCN
 
 | |
5OM9
 
 | | Crystal structure of the human CARBOXYPEPTIDASE A1 in complex with a thiirane mechanism-based inhibitor | | Descriptor: | (2~{R})-4-methyl-2-[(1~{S})-1-sulfanylethyl]pentanoic acid, Carboxypeptidase A1, ZINC ION | | Authors: | Gallego, P, Granados, C, Fernandez, D, Pallares, I, Covaleda, G, Aviles, F.X, Vendrell, J, Reverter, D. | | Deposit date: | 2017-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Mechanism-Based Inactivators for Human Pancreatic Carboxypeptidase A from a Focused Synthetic Library.
ACS Med Chem Lett, 8, 2017
|
|
3NS7
 
 | |
6I6Z
 
 | |
4UEE
 
 | | Crystal structure of the human CARBOXYPEPTIDASE A1 in complex with the PHOSPHINIC INHIBITOR Acetyl-Leu-Ala-Y(PO2CH2)-homoPhe-OH | | Descriptor: | (2S)-2-{[(R)-{(1R)-1-[(N-acetyl-L-leucyl)amino]ethyl}(hydroxy)phosphoryl]methyl}-4-phenylbutanoic acid, CARBOXYPEPTIDASE A1, ZINC ION | | Authors: | Gallego, P, Covaleda, G, Costenaro, L, Devel, L, Dive, V, Aviles, F.X, Reverter, D. | | Deposit date: | 2014-12-17 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of the Human Carboxypeptidase A1 in Complex with Phosphinic Inhibitors
To be Published
|
|
4UEZ
 
 | | Crystal structure of the human CARBOXYPEPTIDASE A1 in complex with the PHOSPHINIC INHIBITOR Acetyl-Leu-Phe-Y(PO2CH2)-Phe-OH | | Descriptor: | (2S)-3-[(R)-{(1R)-1-[(N-acetyl-L-leucyl)amino]-2-phenylethyl}(hydroxy)phosphoryl]-2-benzylpropanoic acid, HUMAN CARBOXYPEPTIDASE A1, ZINC ION | | Authors: | Gallego, P, Covaleda, G, Costenaro, L, Devel, L, Dive, V, Aviles, F.X, Reverter, D. | | Deposit date: | 2014-12-22 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal Structure of the Human Carboxypeptidase A1 in Complex with Phosphinic Inhibitors
To be Published
|
|
6SXJ
 
 | |
6XXQ
 
 | |
6XZ0
 
 | |
6FZB
 
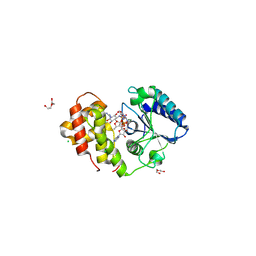 | | AadA in complex with ATP, magnesium and streptomycin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kanchugal P, S, Selmer, M. | | Deposit date: | 2018-03-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural mechanism of AadA, a dual-specificity aminoglycoside adenylyltransferase fromSalmonella enterica.
J. Biol. Chem., 293, 2018
|
|
5AQ0
 
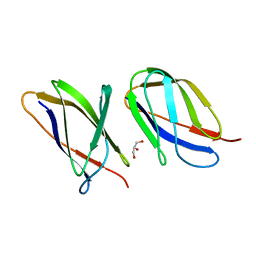 | | The structure of the Transthyretin-like domain of the first catalytic domain of the HUMAN Carboxypeptidase D | | Descriptor: | CARBOXYPEPTIDASE D, GLYCEROL | | Authors: | Gallego, P, Garcia-Pardo, J, Lorenzo, J, Aviles, F.X, Ventura, S, Reverter, D. | | Deposit date: | 2015-09-18 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The Structure of the Ttldomain of the Human Carboxypeptidase D
To be Published
|
|
5AEK
 
 | | Crystal structure of the human SENP2 C548S in complex with the human SUMO1 K48M F66W | | Descriptor: | SENTRIN-SPECIFIC PROTEASE 2, SMALL UBIQUITIN-RELATED MODIFIER 1 | | Authors: | Gallego, P, Grana-Montes, R, Espargaro, A, Castillo, V, Torrent, J, Lange, R, Papaleo, E, Lindorff-Larsend, K, Ventura, S, Reverter, D. | | Deposit date: | 2014-12-23 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Stepping Back and Forward on Sumo Folding Evolution
To be Published
|
|
3OWU
 
 | |
3OX9
 
 | |
3OWS
 
 | |
3OXA
 
 | |
5LQG
 
 | |
5LQH
 
 | |
5FW6
 
 | | Structure of human transthyretin mutant A108V | | Descriptor: | TRANSTHYRETIN | | Authors: | Gallego, P, Varejao, N, Santanna, R, Saraiva, M.J, Ventura, S, Reverter, D. | | Deposit date: | 2016-02-12 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Cavity filling mutations at the thyroxine-binding site dramatically increase transthyretin stability and prevent its aggregation.
Sci Rep, 7, 2017
|
|
3OWY
 
 | |
