5WJI
 
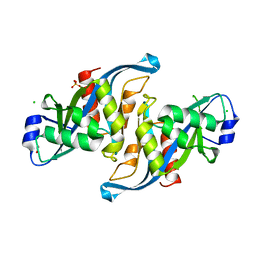 | | Crystal structure of the F61S mutant of HsNUDT16 | | Descriptor: | ACETIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-07-23 | | Release date: | 2018-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
7R63
 
 | |
5WJA
 
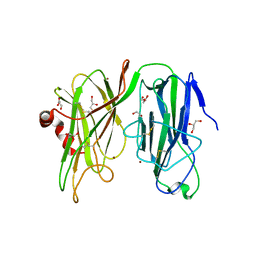 | | Crystal structure of H107A peptidylglycine alpha-hydroxylating monooxygenase (PHM) in complex with citrate | | Descriptor: | CITRATE ANION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-07-21 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
5WKW
 
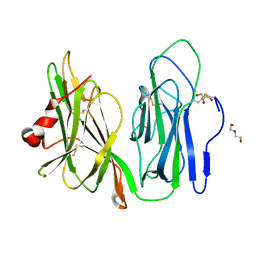 | | Crystal structure of apo wild type peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-07-25 | | Release date: | 2018-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
5WM0
 
 | |
8DVG
 
 | | Structure of KRAS WT(7-16)-HLA-A*03:01 | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, HLA class I histocompatibility antigen, ... | | Authors: | Wright, K.M, Miller, M, Gabelli, S.B. | | Deposit date: | 2022-07-28 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Hydrophobic interactions dominate the recognition of a KRAS G12V neoantigen.
Nat Commun, 14, 2023
|
|
7U1B
 
 | | Crystal structure of EstG in complex with tantalum cluster | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICINE, Beta-lactamase domain-containing protein, ... | | Authors: | Chen, Z, Gabelli, S.B. | | Deposit date: | 2022-02-20 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | EstG is a novel esterase required for cell envelope integrity in Caulobacter.
Curr.Biol., 33, 2023
|
|
7UDA
 
 | | Structure of the EstG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase domain-containing protein, SODIUM ION | | Authors: | Chen, Z, Gabelli, S.B. | | Deposit date: | 2022-03-18 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | EstG is a novel esterase required for cell envelope integrity in Caulobacter.
Curr.Biol., 33, 2023
|
|
7JUL
 
 | | Crystal structure of non phosphorylated PTEN (n-crPTEN-13sp-T1, SDTTDSDPENEG) | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Dempsey, D, Phan, K, Cole, P, Gabelli, S.B. | | Deposit date: | 2020-08-20 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The structural basis of PTEN regulation by multi-site phosphorylation.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4DXJ
 
 | | Crystal structure of Trypanosome cruzi farnesyl diphosphate synthase in complex with [2-(n-propylamino)ethane-1,1-diyl]bisphosphonic acid and Mg2+ | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Aripirala, S, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Design, synthesis, calorimetry, and crystallographic analysis of 2-alkylaminoethyl-1,1-bisphosphonates as inhibitors of Trypanosoma cruzi farnesyl diphosphate synthase.
J.Med.Chem., 55, 2012
|
|
4DWG
 
 | | Crystal structure of Trypanosome cruzi farnesyl diphosphate synthase in complex with [2-(n-heptylamino)ethane-1,1-diyl]bisphosphonic acid and Mg2+ | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Farnesyl pyrophosphate synthase, ... | | Authors: | Aripirala, S, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2012-02-24 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Design, synthesis, calorimetry, and crystallographic analysis of 2-alkylaminoethyl-1,1-bisphosphonates as inhibitors of Trypanosoma cruzi farnesyl diphosphate synthase.
J.Med.Chem., 55, 2012
|
|
4E1E
 
 | | Crystal structure of Trypanosome cruzi farnesyl diphosphate synthase in complex with [2-(n-hexylamino)ethane-1,1-diyl]bisphosphonic acid and Mg2+ | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Aripirala, S, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2012-03-06 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Design, synthesis, calorimetry, and crystallographic analysis of 2-alkylaminoethyl-1,1-bisphosphonates as inhibitors of Trypanosoma cruzi farnesyl diphosphate synthase.
J.Med.Chem., 55, 2012
|
|
4HFQ
 
 | | Crystal structure of UDP-X diphosphatase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Duong-Ly, K.C, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2012-10-05 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A UDP-X diphosphatase from Streptococcus pneumoniae hydrolyzes precursors of peptidoglycan biosynthesis.
Plos One, 8, 2013
|
|
6ALA
 
 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) in complex with citrate | | Descriptor: | CITRATE ANION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-07 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6AN3
 
 | | Crystal structure of H172A-peptidylglycine alpha-hydroxylating monooxygenase (PHM) mutant soaked with peptide (no CuH bound, no peptide bound) | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-11 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6AY0
 
 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) soaked with peptide | | Descriptor: | COPPER (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-09-07 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6AMP
 
 | | Crystal structure of H172A PHM (CuH absent, CuM present) | | Descriptor: | COPPER (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-10 | | Release date: | 2018-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6ALV
 
 | | Crystal structure of H107A-peptidylglycine alpha-hydroxylating monooxygenase (PHM) mutant (no CuH bound) | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-08 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6AO6
 
 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | Descriptor: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-15 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
8E0P
 
 | | Crystal structure of mouse APCDD1 in fusion with engineered MBP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, CHLORIDE ION, ... | | Authors: | Hsieh, F.L, Chang, T.H, Gabelli, S.B, Nathans, J. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of WNT inhibitor adenomatosis polyposis coli down-regulated 1 (APCDD1), a cell-surface lipid-binding protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8E0W
 
 | | Crystal structure of mouse APCDD1 in P1 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hsieh, F.L, Chang, T.H, Gabelli, S.B, Nathans, J. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of WNT inhibitor adenomatosis polyposis coli down-regulated 1 (APCDD1), a cell-surface lipid-binding protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8E0R
 
 | | Crystal structure of mouse APCDD1 in P21 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein APCDD1 | | Authors: | Hsieh, F.L, Chang, T.H, Gabelli, S.B, Nathans, J. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of WNT inhibitor adenomatosis polyposis coli down-regulated 1 (APCDD1), a cell-surface lipid-binding protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FBY
 
 | |
8FCF
 
 | | Crystal structure of PLVAP CC1 in I212121 space group | | Descriptor: | Plasmalemma vesicle-associated protein | | Authors: | Chang, T.H, Hsieh, F.L, Gu, X, Kavran, J, Gabelli, S.B, Nathans, J. | | Deposit date: | 2022-12-01 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into plasmalemma vesicle-associated protein (PLVAP): Implications for vascular endothelial diaphragms and fenestrae.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3NPE
 
 | | Structure of VP14 in complex with oxygen | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 9-cis-epoxycarotenoid dioxygenase 1, chloroplastic, ... | | Authors: | Messing, S.A, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2010-06-28 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid.
Plant Cell, 22, 2010
|
|
