1Y1X
 
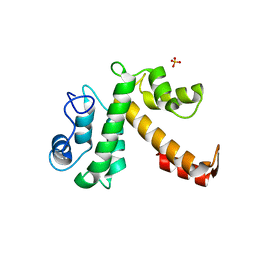 | |
1XQ7
 
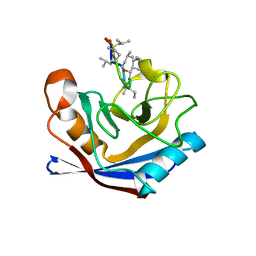 | |
1XTD
 
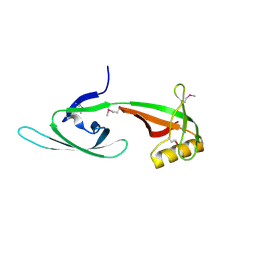 | |
1XTP
 
 | |
1ZLK
 
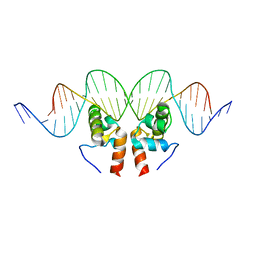 | | Crystal Structure of the Mycobacterium tuberculosis Hypoxic Response Regulator DosR C-terminal Domain-DNA Complex | | Descriptor: | 5'-D(*CP*GP*TP*GP*GP*CP*CP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*AP*CP*TP*TP*TP*AP*GP*TP*CP*CP*CP*CP*AP*AP*AP*GP*CP*GP*CP*GP*GP*GP*CP*CP*AP*T)-3', 5'-D(*GP*GP*CP*CP*CP*GP*CP*GP*CP*TP*TP*TP*GP*GP*GP*GP*AP*CP*TP*AP*AP*AP*GP*TP*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*GP*GP*CP*CP*AP*CP*GP*AP*T)-3', Dormancy Survival Regulator | | Authors: | Wisedchaisri, G, Wu, M, Rice, A.E, Roberts, D.M, Sherman, D.R, Hol, W.G.J. | | Deposit date: | 2005-05-06 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Mycobacterium tuberculosis DosR and DosR-DNA complex involved in gene activation during adaptation to hypoxic latency.
J.Mol.Biol., 354, 2005
|
|
1Z00
 
 | | Solution structure of the C-terminal domain of ERCC1 complexed with the C-terminal domain of XPF | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Tripsianes, K, Folkers, G, Ab, E, Das, D, Odijk, H, Jaspers, N.G.J, Hoeijmakers, J.H.J, Kaptein, R, Boelens, R. | | Deposit date: | 2005-03-01 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Human ERCC1/XPF Interaction Domains Reveals a Complementary Role for the Two Proteins in Nucleotide Excision Repair
Structure, 13, 2005
|
|
1Z2N
 
 | | Inositol 1,3,4-trisphosphate 5/6-kinase complexed Mg2+/ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, inositol 1,3,4-trisphosphate 5/6-kinase | | Authors: | Miller, G.J, Wilson, M.P, Majerus, P.W, Hurley, J.H. | | Deposit date: | 2005-03-08 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase.
Mol.Cell, 18, 2005
|
|
1UHV
 
 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | 1,5-anhydro-2-deoxy-2-fluoro-D-xylitol, Beta-xylosidase | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-11 | | Release date: | 2003-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
1UP3
 
 | | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with METHYL-CELLOBIOSYL-4-DEOXY-4-THIO-BETA-D-CELLOBIOSIDE at 1.6 angstrom | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, PUTATIVE CELLULASE CEL6, SULFATE ION, ... | | Authors: | Varrot, A, Leydier, S, Pell, G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-09-26 | | Release date: | 2004-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mycobacterium Tuberculosis Strains Possess Functional Cellulases.
J.Biol.Chem., 280, 2005
|
|
1UWR
 
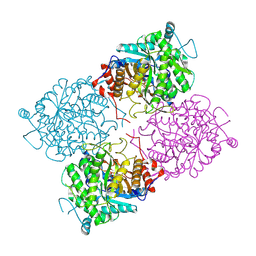 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with 2-deoxy-2-fluoro-galactose | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-galactopyranose, ACETATE ION, BETA-GALACTOSIDASE | | Authors: | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Studies of the Beta-Glycosidase from Sulfolobus Solfataricus in Complex with Covalently and Noncovalently Bound Inhibitors.
Biochemistry, 43, 2004
|
|
1XJS
 
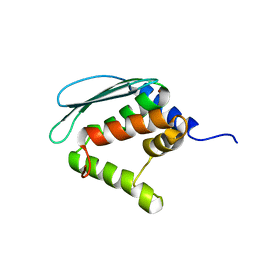 | | Solution structure of Iron-Sulfur cluster assembly protein IscU from Bacillus subtilis, with Zinc bound at the active site. Northeast Structural Genomics Consortium Target SR17 | | Descriptor: | NifU-like protein, ZINC ION | | Authors: | Kornhaber, G.J, Swapna, G.V.T, Ramelot, T.A, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-24 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Iron-Sulfur cluster assembly protein IscU
from Bacillus subtilis, with Zinc bound at the active site.
Northeast Structural Genomics Consortium Target SR17.
To be Published
|
|
7ONS
 
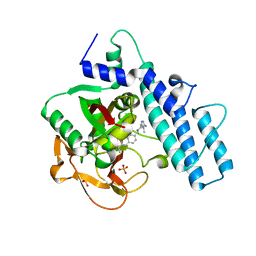 | | PARP1 catalytic domain in complex with isoquinolone-based inhibitor (compound 16) | | Descriptor: | 7-[[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]methyl]-3-ethyl-1~{H}-quinolin-2-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
7ONR
 
 | | PARP1 catalytic domain in complex with 8-chloroquinazolinone-based inhibitor (compound 9) | | Descriptor: | 8-chloranyl-2-[3-[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]propyl]-3~{H}-quinazolin-4-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
7ONT
 
 | | PARP1 catalytic domain in complex with a selective pyridine carboxamide-based inhibitor (compound 22) | | Descriptor: | 5-[4-[(3-ethyl-2-oxidanylidene-1~{H}-quinolin-7-yl)methyl]piperazin-1-yl]-~{N}-methyl-pyridine-2-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M.J, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
1RK4
 
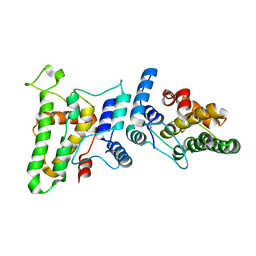 | | Crystal Structure of a Soluble Dimeric Form of Oxidised CLIC1 | | Descriptor: | CHLORIDE INTRACELLULAR CHANNEL PROTEIN 1 | | Authors: | Littler, D.R, Harrop, S.J, Fairlie, W.D, Brown, L.J, Pankhurst, G.J, Pankhurst, S, DeMaere, M.Z, Campbell, T.J, Bauskin, A.R, Tonini, R, Mazzanti, M, Breit, S.N, Curmi, P.M. | | Deposit date: | 2003-11-20 | | Release date: | 2003-12-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | The Intracellular Chloride Ion Channel Protein CLIC1 Undergoes a Redox-controlled Structural Transition
J.Biol.Chem., 279, 2004
|
|
1RHD
 
 | |
1Z2P
 
 | | Inositol 1,3,4-trisphosphate 5/6-Kinase in complex with Mg2+/AMP-PCP/Ins(1,3,4)P3 | | Descriptor: | (1S,3S,4S)-1,3,4-TRIPHOSPHO-MYO-INOSITOL, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Miller, G.J, Wilson, M.P, Majerus, P.W, Hurley, J.H. | | Deposit date: | 2005-03-08 | | Release date: | 2005-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase.
Mol.Cell, 18, 2005
|
|
1YJ8
 
 | |
1UOZ
 
 | | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with thiocellopentaose at 1.1 angstrom | | Descriptor: | 4-thio-beta-D-glucopyranose, GLYCEROL, PUTATIVE CELLULASE, ... | | Authors: | Varrot, A, Leydier, S, Pell, G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-09-26 | | Release date: | 2004-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mycobacterium Tuberculosis Strains Possess Functional Cellulases.
J.Biol.Chem., 280, 2005
|
|
1US3
 
 | | Native xylanase10C from Cellvibrio japonicus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-BETA-1,4-XYLANASE PRECURSOR, GLYCEROL, ... | | Authors: | Pell, G, Szabo, L, Charnock, S.J, Xie, H, Gloster, T.M, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-11-17 | | Release date: | 2003-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Analysis of Cellvibrio Japonicus Xylanase 10C: How Variation in Substrate-Binding Cleft Influences the Catalytic Profile of Family Gh-10 Xylanases
J.Biol.Chem., 279, 2004
|
|
1UQZ
 
 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with 4-O-methyl glucuronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, CHLORIDE ION, ENDOXYLANASE, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-24 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
1UP7
 
 | | Structure of the 6-phospho-beta glucosidase from Thermotoga maritima at 2.4 Angstrom resolution in the tetragonal form with NAD and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-PHOSPHO-BETA-GLUCOSIDASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Varrot, A, Yip, V.L, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-09-29 | | Release date: | 2004-11-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nad+ and Metal-Ion Dependent Hydrolysis by Family 4 Glycosidases: Structural Insight Into Specificity for Phospho-Beta-D-Glucosides
J.Mol.Biol., 346, 2005
|
|
1V0K
 
 | | Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 5.8 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, PIPERIDINE-3,4,5-TRIOL, beta-D-xylopyranose | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki S, J, Withers, G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
1V0M
 
 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 7.5 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, PIPERIDINE-3,4,5-TRIOL, ... | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
1UWT
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with D-galactohydroximo-1,5-lactam | | Descriptor: | (2E,3R,4R,5R,6S)-3,4,5-TRIHYDROXY-6-(HYDROXYMETHYL)-2-PIPERIDINONE, ACETATE ION, BETA-GALACTOSIDASE | | Authors: | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of the beta-glycosidase from Sulfolobus solfataricus in complex with covalently and noncovalently bound inhibitors.
Biochemistry, 43, 2004
|
|
