3OV8
 
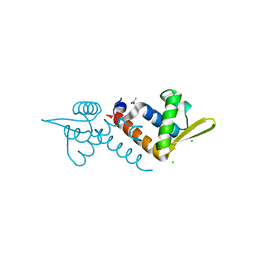 | | Crystal structure of AF1382 from Archaeoglobus fulgidus, High resolution | | Descriptor: | ACETATE ION, CHLORIDE ION, Protein AF_1382 | | Authors: | Zhu, J.-Y, Zhao, M, Fu, Z.-Q, Yang, H, Chang, J, Hao, X, Chen, L, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2010-09-16 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8501 Å) | | Cite: | Structure of the Archaeoglobus fulgidus orphan ORF AF1382 determined by sulfur SAD from a moderately diffracting crystal.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6WIV
 
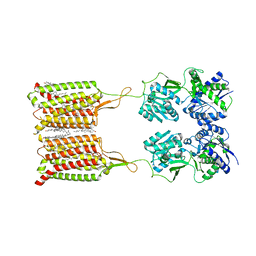 | | Structure of human GABA(B) receptor in an inactive state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-{[(9Z)-octadec-9-enoyl]oxy}propyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Fu, Z, Frangaj, A, Liu, J, Mosyak, L, Shen, T, Slavkovich, V.N, Ray, K.M, Taura, J, Cao, B, Geng, Y, Zuo, H, Kou, Y, Grassucci, R, Chen, S, Liu, Z, Lin, X, Williams, J.P, Rice, W.J, Eng, E.T, Huang, R.K, Soni, R.K, Kloss, B, Yu, Z, Javitch, J.A, Hendrickson, W.A, Slesinger, P.A, Quick, M, Graziano, J, Yu, H, Fiehn, O, Clarke, O.B, Frank, J, Fan, Q.R. | | Deposit date: | 2020-04-10 | | Release date: | 2020-07-01 | | Last modified: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human GABABreceptor in an inactive state.
Nature, 584, 2020
|
|
3O3K
 
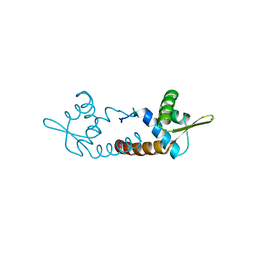 | | Crystal structure of AF1382 from Archaeoglobus fulgidus | | Descriptor: | Uncharacterized protein AF_1382 | | Authors: | Zhu, J.-Y, Zhao, M, Fu, Z.-Q, Yang, H, Chang, J, Xu, H, Chen, L, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2010-07-24 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Archaeoglobus fulgidus orphan ORF AF1382 determined by sulfur SAD from a moderately diffracting crystal.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1A8M
 
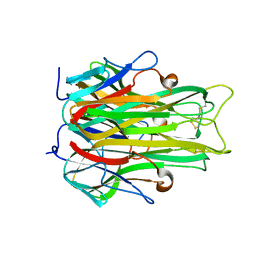 | | TUMOR NECROSIS FACTOR ALPHA, R31D MUTANT | | Descriptor: | TUMOR NECROSIS FACTOR ALPHA | | Authors: | Reed, C, Fu, Z.-Q, Wu, J, Xue, Y.-N, Harrison, R.W, Chen, M.-J, Weber, I.T. | | Deposit date: | 1998-03-27 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of TNF-alpha mutant R31D with greater affinity for receptor R1 compared with R2.
Protein Eng., 10, 1997
|
|
6WEK
 
 | | Structure of cGMP-bound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLIC GUANOSINE MONOPHOSPHATE, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WEL
 
 | | Structure of cGMP-unbound F403V/V407A mutant TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WEJ
 
 | | Structure of cGMP-unbound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2EIS
 
 | | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8 | | Descriptor: | COENZYME A, Hypothetical protein TTHB207 | | Authors: | Kamitori, S, Yoshida, H, Satoh, S, Iino, H, Ebihara, A, Chen, L, Fu, Z.-Q, Chrzas, J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-13 | | Release date: | 2008-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8
To be Published
|
|
2EJ5
 
 | | Crystal structure of GK2038 protein (enoyl-CoA hydratase subunit II) from Geobacillus kaustophilus | | Descriptor: | Enoyl-CoA hydratase subunit II | | Authors: | Okazaki, N, Agari, Y, Ebihara, A, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Kuramitsu, S, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-15 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GK2038 protein (enoyl-CoA hydratase subunit II) from Geobacillus kaustophilus
To be Published
|
|
1SIR
 
 | | The Crystal Structure and Mechanism of Human Glutaryl-CoA Dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, S-4-NITROBUTYRYL-COA | | Authors: | Wang, M, Fu, Z, Paschke, R, Goodman, S.L, Frerman, F.E, Kim, J.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Human Glutaryl-CoA Dehydrogenase with and without an Alternate Substrate: Structural Bases of Dehydrogenation and Decarboxylation Reactions
Biochemistry, 43, 2004
|
|
1Y80
 
 | | Structure of a corrinoid (factor IIIm)-binding protein from Moorella thermoacetica | | Descriptor: | CO-5-METHOXYBENZIMIDAZOLYLCOBAMIDE, Predicted cobalamin binding protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Z.-J, Fu, Z.-Q, Tempel, W, Das, A, Habel, J, Zhou, W, Chang, J, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Ljungdahl, L, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a corrinoid (factor IIIm)-binding protein from Moorella thermoacetica
To be published
|
|
2I0X
 
 | | Hypothetical protein PF1117 from Pyrococcus furiosus | | Descriptor: | Hypothetical protein PF1117 | | Authors: | Chen, L.Q, Fu, Z.-Q, Liu, Z.-J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-08-11 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Hypothetical Protein Pf1117 from Pyrococcus furiosus
To be Published
|
|
1SIQ
 
 | | The Crystal Structure and Mechanism of Human Glutaryl-CoA Dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase | | Authors: | Wang, M, Fu, Z, Paschke, R, Goodman, S, Frerman, F.E, Kim, J.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Human Glutaryl-CoA Dehydrogenase with and without an Alternate Substrate: Structural Bases of Dehydrogenation and Decarboxylation Reactions
Biochemistry, 43, 2004
|
|
2YX5
 
 | | Crystal Structure of Methanocaldococcus jannaschii PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway | | Descriptor: | UPF0062 protein MJ1593 | | Authors: | Kanagawa, M, Baba, S, Agari, Y, Chen, L.Q, Fu, Z.-Q, Chrzas, J, Wang, B.C, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Methanocaldococcus jannaschii PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway
To be Published
|
|
3QK9
 
 | | Yeast Tim44 C-terminal domain complexed with Cymal-3 | | Descriptor: | CHLORIDE ION, Mitochondrial import inner membrane translocase subunit TIM44 | | Authors: | Cui, W, Josyula, R, Fu, Z, Sha, B. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Membrane Binding Mechanism of Yeast Mitochondrial Peripheral Membrane Protein TIM44.
Protein Pept.Lett., 18, 2011
|
|
2YYO
 
 | | Crystal structure of human SPRY domain | | Descriptor: | SPRY domain-containing protein 3 | | Authors: | Kishishita, S, Uchikubo-Kamo, T, Murayama, K, Terada, T, Chen, L, Fu, Z.Q, Chrzas, J, Shirouzu, M, Wang, B.C, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-30 | | Release date: | 2008-05-06 | | Last modified: | 2020-09-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human SPRY domain
To be Published
|
|
7URI
 
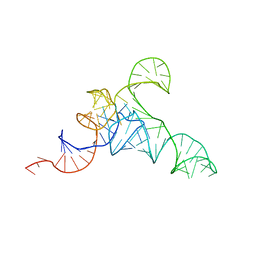 | | allo-tRNAUTu1A in the A site of the E. coli ribosome | | Descriptor: | Allo-tRNAUTu1A | | Authors: | Zhang, J, Prabhakar, A, Krahn, N, Vargas-Rodriguez, O, Krupkin, M, Fu, Z, Acosta-Reyes, F.J, Ge, X, Choi, J, Crnkovic, A, Ehrenberg, M, Viani Puglisi, E, Soll, D, Puglisi, J. | | Deposit date: | 2022-04-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Uncovering translation roadblocks during the development of a synthetic tRNA.
Nucleic Acids Res., 50, 2022
|
|
7UR5
 
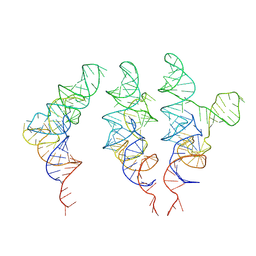 | | allo-tRNAUTu1 in the A, P, and E sites of the E. coli ribosome | | Descriptor: | allo-tRNAUTu1 | | Authors: | Zhang, J, Krahn, N, Prabhakar, A, Vargas-Rodriguez, O, Krupkin, M, Fu, Z, Acosta-Reyes, F.J, Ge, X, Choi, J, Crnkovic, A, Ehrenberg, M, Viani Puglisi, E, Soll, D, Puglisi, J. | | Deposit date: | 2022-04-21 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Uncovering translation roadblocks during the development of a synthetic tRNA.
Nucleic Acids Res., 50, 2022
|
|
7URM
 
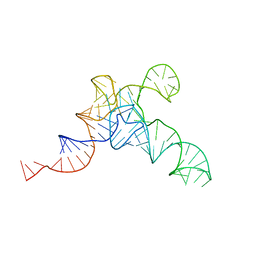 | | allo-tRNAUTu1A in the P site of the E. coli ribosome | | Descriptor: | allo-tRNAUTU1A | | Authors: | Zhang, J, Prabhakar, A, Krahn, N, Vargas-Rodriguez, O, Krupkin, M, Fu, Z, Acosta-Reyes, F.J, Ge, X, Choi, J, Crnkovic, A, Ehrenberg, M, Viani Puglisi, E, Soll, D, Puglisi, J. | | Deposit date: | 2022-04-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Uncovering translation roadblocks during the development of a synthetic tRNA.
Nucleic Acids Res., 50, 2022
|
|
1Z2M
 
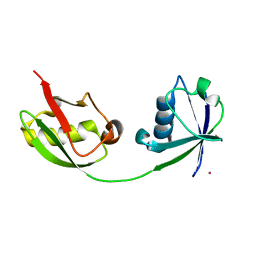 | | Crystal Structure of ISG15, the Interferon-Induced Ubiquitin Cross Reactive Protein | | Descriptor: | OSMIUM 4+ ION, interferon, alpha-inducible protein (clone IFI-15K) | | Authors: | Narasimhan, J, Wang, M, Fu, Z, Klein, J.M, Haas, A.L, Kim, J.J. | | Deposit date: | 2005-03-08 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Interferon-induced Ubiquitin-like Protein ISG15.
J.Biol.Chem., 280, 2005
|
|
2YZ8
 
 | | Crystal structure of the 32th Ig-like domain of human obscurin (KIAA1556) | | Descriptor: | Obscurin | | Authors: | Saijo, S, Ohsawa, N, Nishino, A, Kishishita, S, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-04 | | Release date: | 2008-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the 32th Ig-like domain of human obscurin (KIAA1556)
To be Published
|
|
2Z0M
 
 | | Crystal structure of hypothetical ATP-dependent RNA helicase from Sulfolobus tokodaii | | Descriptor: | 337aa long hypothetical ATP-dependent RNA helicase deaD | | Authors: | Nakagawa, N, Kusano, S, Shirouzu, M, Chen, L, Fu, Z.-Q, Chrzas, J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of hypothetical ATP-dependent RNA helicase from Sulfolobus tokodaii
To be Published
|
|
2ZBV
 
 | | Crystal structure of uncharacterized conserved protein from Thermotoga maritima | | Descriptor: | ADENOSINE, Uncharacterized conserved protein | | Authors: | Ebihara, A, Fujimoto, Y, Kagawa, W, Fujikawa, N, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of uncharacterized conserved protein from Thermotoga maritima
To be Published
|
|
6VX7
 
 | | bestrophin-2 Ca2+-bound state (5 mM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX8
 
 | | bestrophin-2 Ca2+- unbound state 2 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
