2WGN
 
 | | Pseudomonas aeruginosa ICP | | Descriptor: | INHIBITOR OF CYSTEINE PEPTIDASE COMPND 3 | | Authors: | Fu, Y, Smith, B.O. | | Deposit date: | 2009-04-21 | | Release date: | 2010-07-07 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of Pseudomonas Aeruginosa Icp
Ph D Thesis, 2009
|
|
4F2E
 
 | |
4F2F
 
 | | Crystal structure of the metal binding domain (MBD) of the Streptococcus pneumoniae D39 Cu(I) exporting P-type ATPase CopA with Cu(I) | | Descriptor: | CHLORIDE ION, COPPER (I) ION, Cation-transporting ATPase, ... | | Authors: | Fu, Y, Dann III, C.E, Giedroc, D.P. | | Deposit date: | 2012-05-07 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A new structural paradigm in copper resistance in Streptococcus pneumoniae.
Nat.Chem.Biol., 9, 2013
|
|
1XI2
 
 | | Quinone Reductase 2 in Complex with Cancer Prodrug CB1954 | | Descriptor: | 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, NRH dehydrogenase [quinone] 2, ... | | Authors: | Fu, Y, Buryanovskyy, L, Zhang, Z. | | Deposit date: | 2004-09-21 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of quinone reductase 2 in complex with cancer prodrug CB1954
Biochem.Biophys.Res.Commun., 336, 2005
|
|
2QMZ
 
 | |
2MRY
 
 | |
7YPQ
 
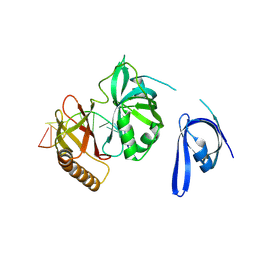 | | Cryo-EM structure of one baculovirus LEF-3 molecule in complex with ssDNA | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Lef3 | | Authors: | Fu, Y, Rao, G, Yin, J, Li, Z, Cao, S. | | Deposit date: | 2022-08-04 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural transitions during the cooperative assembly of baculovirus single-stranded DNA-binding protein on ssDNA.
Nucleic Acids Res., 50, 2022
|
|
4ME3
 
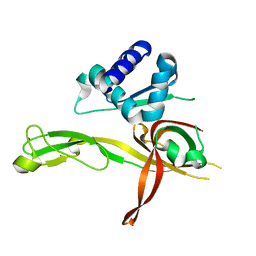 | | 1.8 Angstrom Crystal Structure of the N-terminal Domain of an Archaeal MCM | | Descriptor: | DNA replication licensing factor MCM related protein, ZINC ION | | Authors: | Fu, Y, Slaymaker, I.M, Wang, G, Chen, X.S. | | Deposit date: | 2013-08-24 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | The 1.8- angstrom Crystal Structure of the N-Terminal Domain of an Archaeal MCM as a Right-Handed Filament.
J.Mol.Biol., 426, 2014
|
|
9J03
 
 | | Cyro-EM Structure of Human TLR4/MD-2/DLAM1 Complex | | Descriptor: | (3R)-3-(dodecanoyloxy)tetradecanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fu, Y, Kim, H, Zamyatina, A, Kim, H.M. | | Deposit date: | 2024-08-02 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insight into TLR4/MD-2 activation by synthetic LPS mimetics with distinct binding modes.
Nat Commun, 16, 2025
|
|
5YSS
 
 | |
5YSZ
 
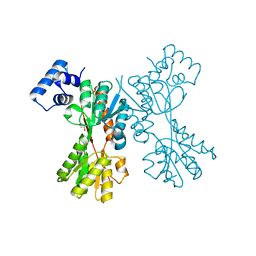 | | transcriptional regulator CelR-cellobiose complex | | Descriptor: | GLYCEROL, Transcriptional regulator, LacI family, ... | | Authors: | Fu, Y, Yeom, S.Y, Lee, D.H, Lee, S.G. | | Deposit date: | 2017-11-16 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Structural and functional analyses of the cellulase transcription regulator CelR
FEBS Lett., 592, 2018
|
|
4R3Z
 
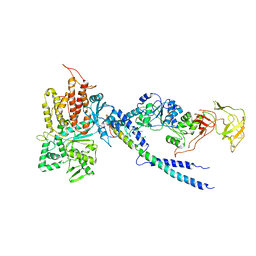 | | Crystal structure of human ArgRS-GlnRS-AIMP1 complex | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 1, Arginine--tRNA ligase, cytoplasmic, ... | | Authors: | Fu, Y, Kim, Y, Cho, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.033 Å) | | Cite: | Structure of the ArgRS-GlnRS-AIMP1 complex and its implications for mammalian translation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8WO1
 
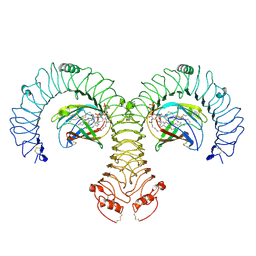 | | Cryo-EM Structure of Human TLR4/MD-2/DLAM5 Complex | | Descriptor: | (3R)-3-(dodecanoyloxy)tetradecanoic acid, (3R)-3-(tetradecanoyloxy)tetradecanoic acid, (3~{S})-3-decanoyloxytetradecanoic acid, ... | | Authors: | Fu, Y, Kim, H, Zamyatina, A, Kim, H.M. | | Deposit date: | 2023-10-06 | | Release date: | 2024-10-09 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Structural insight into TLR4/MD-2 activation by synthetic LPS mimetics with distinct binding modes.
Nat Commun, 16, 2025
|
|
8WSA
 
 | | Cryo-EM Structure of Mouse TLR4/MD-2/DLAM5 Complex | | Descriptor: | (3R)-3-(dodecanoyloxy)tetradecanoic acid, (3R)-3-(tetradecanoyloxy)tetradecanoic acid, (3~{S})-3-decanoyloxytetradecanoic acid, ... | | Authors: | Fu, Y, Kim, H, Zamyatina, A, Kim, H.M. | | Deposit date: | 2023-10-17 | | Release date: | 2024-10-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into TLR4/MD-2 activation by synthetic LPS mimetics with distinct binding modes.
Nat Commun, 16, 2025
|
|
8WQT
 
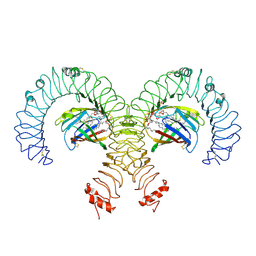 | | Cryo-EM Structure of Mouse TLR4/MD-2/DLAM1 complex | | Descriptor: | (3R)-3-(dodecanoyloxy)tetradecanoic acid, (3R)-3-(tetradecanoyloxy)tetradecanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fu, Y, Kim, H, Zamyatina, A, Kim, H.M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-16 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insight into TLR4/MD-2 activation by synthetic LPS mimetics with distinct binding modes.
Nat Commun, 16, 2025
|
|
8WRY
 
 | | Cryo-EM Structure of Mouse TLR4/MD-2/DLAM3 Complex | | Descriptor: | (3R)-3-(dodecanoyloxy)tetradecanoic acid, (3R)-3-(tetradecanoyloxy)tetradecanoic acid, 2-(hydroxymethyl)-5-methoxy-3,6-bis(oxidanyl)pyran-4-one, ... | | Authors: | Fu, Y, Kim, H, Zamyatina, A, Kim, H.M. | | Deposit date: | 2023-10-16 | | Release date: | 2024-10-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insight into TLR4/MD-2 activation by synthetic LPS mimetics with distinct binding modes.
Nat Commun, 16, 2025
|
|
8WTA
 
 | | Cryo-EM Structure of Human TLR4/MD-2/DLAM3 Complex | | Descriptor: | (3R)-3-(dodecanoyloxy)tetradecanoic acid, (3R)-3-(tetradecanoyloxy)tetradecanoic acid, 2-(hydroxymethyl)-5-methoxy-3,6-bis(oxidanyl)pyran-4-one, ... | | Authors: | Fu, Y, Kim, H, Zamyatina, A, Kim, H.M. | | Deposit date: | 2023-10-18 | | Release date: | 2024-10-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into TLR4/MD-2 activation by synthetic LPS mimetics with distinct binding modes.
Nat Commun, 16, 2025
|
|
4FDG
 
 | | Crystal Structure of an Archaeal MCM Filament | | Descriptor: | Minichromosome maintenance protein MCM, ZINC ION | | Authors: | Slaymaker, I.M, Fu, Y, Brewster, A.B, Chen, X.S. | | Deposit date: | 2012-05-28 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Mini-chromosome maintenance complexes form a filament to remodel DNA structure and topology.
Nucleic Acids Res., 41, 2013
|
|
6ON1
 
 | | A resting state structure of L-DOPA dioxygenase from Streptomyces sclerotialus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, L-DOPA dioxygenase | | Authors: | Wang, Y, Shin, I, Fu, Y, Colabroy, K, Liu, A. | | Deposit date: | 2019-04-19 | | Release date: | 2019-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Crystal Structures of L-DOPA Dioxygenase fromStreptomyces sclerotialus.
Biochemistry, 58, 2019
|
|
7XN4
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with NAD+ | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
7XN5
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Arginine ADP-riboxanase CopC, Calmodulin-1, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
5SZI
 
 | | Structure of human Rab8a in complex with the bMERB domain of Mical-cL | | Descriptor: | MAGNESIUM ION, MICAL C-terminal-like protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Rai, A, Oprisko, A, Campos, J, Fu, Y, Friese, T, Itzen, A, Goody, R.S, Mueller, M.P, Gazdag, E.M. | | Deposit date: | 2016-08-14 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | bMERB domains are bivalent Rab8 family effectors evolved by gene duplication.
Elife, 5, 2016
|
|
5SZJ
 
 | | Structure of human Rab10 in complex with the bMERB domain of Mical-cL | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, MICAL C-terminal-like protein, ... | | Authors: | Rai, A, Oprisko, A, Campos, J, Fu, Y, Friese, T, Itzen, A, Goody, R.S, Mueller, M.P, Gazdag, E.M. | | Deposit date: | 2016-08-14 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | bMERB domains are bivalent Rab8 family effectors evolved by gene duplication.
Elife, 5, 2016
|
|
5SZH
 
 | | Structure of human Rab1b in complex with the bMERB domain of Mical-cL | | Descriptor: | MAGNESIUM ION, MICAL C-terminal-like protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Rai, A, Oprisko, A, Campos, J, Fu, Y, Friese, T, Goody, R.S, Mueller, M.P, Gazdag, E.M. | | Deposit date: | 2016-08-14 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | bMERB domains are bivalent Rab8 family effectors evolved by gene duplication.
Elife, 5, 2016
|
|
5SZG
 
 | | Structure of the bMERB domain of Mical-3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protein-methionine sulfoxide oxidase MICAL3 | | Authors: | Rai, A, Oprisko, A, Campos, J, Fu, Y, Friese, T, Itzen, A, Goody, R.S, Gazdag, E.M, Mueller, M.P. | | Deposit date: | 2016-08-14 | | Release date: | 2016-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | bMERB domains are bivalent Rab8 family effectors evolved by gene duplication.
Elife, 5, 2016
|
|
