1HU0
 
 | | CRYSTAL STRUCTURE OF AN HOGG1-DNA BOROHYDRIDE TRAPPED INTERMEDIATE COMPLEX | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 8-OXOGUANINE, ... | | Authors: | Fromme, J.C, Bruner, S.D, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2001-01-03 | | Release date: | 2003-02-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Product-Assisted Catalysis in base-excision DNA Repair
Nat.Struct.Biol., 10, 2003
|
|
1VRL
 
 | | MutY adenine glycosylase in complex with DNA and soaked adenine free base | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*T)-3', ADENINE, ... | | Authors: | Fromme, J.C, Banerjee, A, Huang, S.J, Verdine, G.L. | | Deposit date: | 2005-03-08 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase
Nature, 427, 2004
|
|
1R2Z
 
 | | MutM (Fpg) bound to 5,6-dihydrouracil (DHU) containing DNA | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*TP*CP*CP*AP*(DHU)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2003-09-30 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | DNA Lesion Recognition by the Bacterial Repair Enzyme MutM.
J.Biol.Chem., 278, 2003
|
|
1R2Y
 
 | | MutM (Fpg) bound to 8-oxoguanine (oxoG) containing DNA | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*TP*CP*CP*AP*(8OG)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2003-09-30 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | DNA Lesion Recognition by the Bacterial Repair Enzyme MutM.
J.Biol.Chem., 278, 2003
|
|
1RRS
 
 | | MutY adenine glycosylase in complex with DNA containing an abasic site | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*T)-3', CALCIUM ION, ... | | Authors: | Fromme, J.C, Banerjee, A, Huang, S.J, Verdine, G.L. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase
Nature, 427, 2004
|
|
1RRQ
 
 | | MutY adenine glycosylase in complex with DNA containing an A:oxoG pair | | Descriptor: | 5'-D(*TP*GP*TP*CP*CP*AP*AP*GP*TP*CP*T)-3', 5'-D(AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3', CALCIUM ION, ... | | Authors: | Fromme, J.C, Banerjee, A, Huang, S.J, Verdine, G.L. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase
Nature, 427, 2004
|
|
1ORP
 
 | |
1ORN
 
 | |
1LWY
 
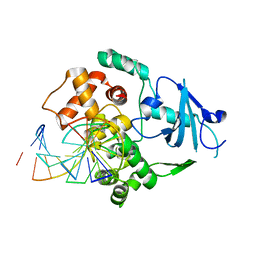 | | hOgg1 Borohydride-Trapped Intermediate without 8-oxoguanine | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 8-OXOGUANINE DNA GLYCOSYLASE | | Authors: | Fromme, J.C, Bruner, S.D, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2002-06-03 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Product-Assisted Catalysis in Base Excision DNA Repair
Nat.Struct.Biol., 10, 2003
|
|
1P59
 
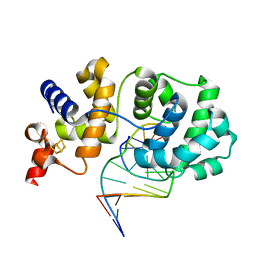 | | Structure of a non-covalent Endonuclease III-DNA Complex | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*GP*(5IU)P*GP*GP*AP*C)-3', 5'-D(TP*GP*(5IU)P*CP*CP*AP*(3DR)P*GP*(5IU)P*CP*T)-3', Endonuclease III, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2003-04-25 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Trapped Endonuclease III-DNA Covalent Intermediate
Embo J., 22, 2003
|
|
1L2D
 
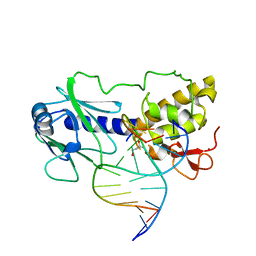 | | MutM (Fpg)-DNA Estranged Guanine Mismatch Recognition Complex | | Descriptor: | 5'-D(*AP*G*GP*TP*AP*GP*AP*CP*GP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-02-20 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM.
Nat.Struct.Biol., 9, 2002
|
|
1L1Z
 
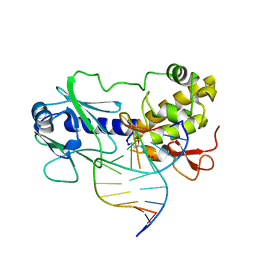 | | MutM (Fpg) Covalent-DNA Intermediate | | Descriptor: | 5'-D(*AP*G*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-02-20 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM.
Nat.Struct.Biol., 9, 2002
|
|
1L2B
 
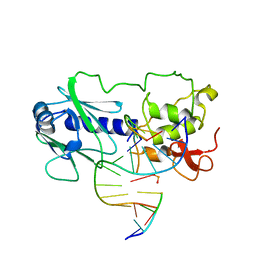 | | MutM (Fpg) DNA End-Product Structure | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*TP*CP*C*(AD2))-3', 5'-D(P*GP*TP*CP*TP*AP*CP*C)-3', ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-02-20 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM.
Nat.Struct.Biol., 9, 2002
|
|
1L2C
 
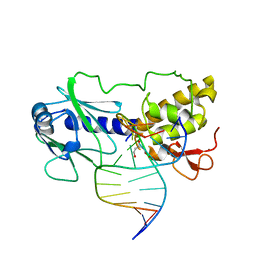 | | MutM (Fpg)-DNA Estranged Thymine Mismatch Recognition Complex | | Descriptor: | 5'-D(*AP*G*GP*TP*AP*GP*AP*CP*TP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-02-20 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM.
Nat.Struct.Biol., 9, 2002
|
|
1L1T
 
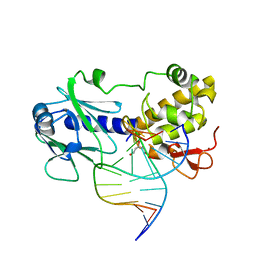 | | MutM (Fpg) Bound to Abasic-Site Containing DNA | | Descriptor: | 5'-D(*AP*G*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-02-19 | | Release date: | 2002-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM.
Nat.Struct.Biol., 9, 2002
|
|
1LWW
 
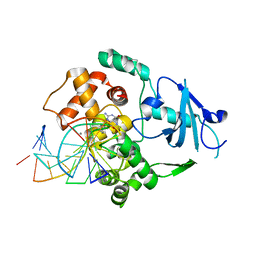 | | Borohydride-trapped hOgg1 Intermediate Structure Co-Crystallized with 8-bromoguanine | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 8-BROMOGUANINE, ... | | Authors: | Fromme, J.C, Bruner, S.D, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2002-06-03 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Product-Assisted Catalysis in Base Excision DNA Repair
Nat.Struct.Biol., 10, 2003
|
|
1LWV
 
 | | Borohydride-trapped hOgg1 Intermediate Structure Co-Crystallized with 8-aminoguanine | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 8-AMINOGUANINE, ... | | Authors: | Fromme, J.C, Bruner, S.D, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2002-06-03 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Product-Assisted Catalysis in Base Excision DNA Repair
Nat.Struct.Biol., 10, 2003
|
|
9AZX
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) Nsp3 macrodomain in complex with NDPr | | Descriptor: | Non-structural protein 3, {(2R,3S,4R,5R)-5-[(8S)-4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl]-5-cyano-3,4-dihydroxyoxolan-2-yl}methyl [(2R,3S,4R,5R)-3,4,5-trihydroxyoxolan-2-yl]methyl dihydrogen diphosphate | | Authors: | Wallace, S.D, Bagde, S.R, Fromme, J.C. | | Deposit date: | 2024-03-11 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | GS-441524-Diphosphate-Ribose Derivatives as Nanomolar Binders and Fluorescence Polarization Tracers for SARS-CoV-2 and Other Viral Macrodomains.
Acs Chem.Biol., 19, 2024
|
|
8CTQ
 
 | | Crystal structure of engineered phospholipase D mutant superPLD 2-48 | | Descriptor: | PHOSPHATE ION, Phospholipase D | | Authors: | Tei, R, Bagde, S.R, Fromme, J.C, Baskin, J.M. | | Deposit date: | 2022-05-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Activity-based directed evolution of a membrane editor in mammalian cells.
Nat.Chem., 15, 2023
|
|
8CTP
 
 | | Crystal structure of engineered phospholipase D mutant superPLD 2-23 | | Descriptor: | PHOSPHATE ION, Phospholipase D | | Authors: | Tei, R, Bagde, S.R, Fromme, J.C, Baskin, J.M. | | Deposit date: | 2022-05-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activity-based directed evolution of a membrane editor in mammalian cells.
Nat.Chem., 15, 2023
|
|
8UCQ
 
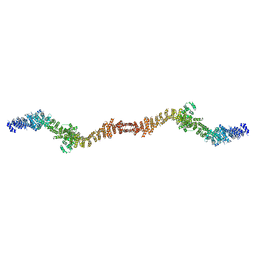 | |
8GIA
 
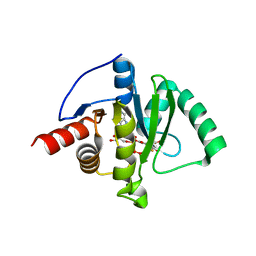 | | Crystal structure of SARS-CoV-2 (Covid-19) Nsp3 macrodomain in complex with TFMU-ADPr | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl [(2R,3S,4R,5R)-3,4-dihydroxy-5-{[2-oxo-4-(trifluoromethyl)-2H-1-benzopyran-7-yl]oxy}oxolan-2-yl]methyl dihydrogen diphosphate | | Authors: | Wallace, S.D, Bagde, S.R, Fromme, J.C. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Fluorescence Polarization Assay for Macrodomains Facilitates the Identification of Potent Inhibitors of the SARS-CoV-2 Macrodomain.
Acs Chem.Biol., 18, 2023
|
|
8DQ0
 
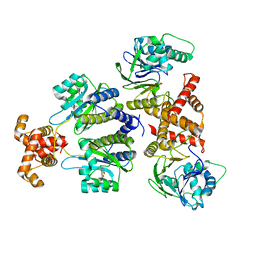 | | Quorum-sensing receptor RhlR bound to PqsE | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, RhlR protein | | Authors: | Paczkowski, J.E, Fromme, J.C, Feathers, J.R. | | Deposit date: | 2022-07-18 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structure of the RhlR-PqsE complex from Pseudomonas aeruginosa reveals mechanistic insights into quorum-sensing gene regulation.
Structure, 30, 2022
|
|
8DQ1
 
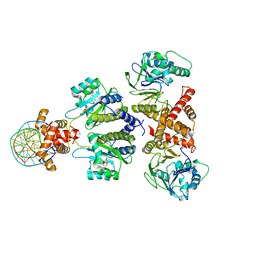 | | Quorum-sensing receptor RhlR bound to PqsE | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, DNA (5'-D(*AP*CP*CP*TP*GP*CP*CP*AP*GP*AP*CP*TP*GP*CP*AP*CP*AP*G)-3'), ... | | Authors: | Paczkowski, J.E, Fromme, J.C, Feathers, J.R. | | Deposit date: | 2022-07-18 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the RhlR-PqsE complex from Pseudomonas aeruginosa reveals mechanistic insights into quorum-sensing gene regulation.
Structure, 30, 2022
|
|
7URO
 
 | |
