2NT9
 
 | | Crystal structure of pectin methylesterase D178A mutant in complex with hexasaccharide IV | | Descriptor: | Pectinesterase A, alpha-D-galactopyranuronic acid-(1-4)-methyl alpha-D-galactopyranuronate-(1-4)-methyl alpha-D-galactopyranuronate-(1-4)-methyl alpha-D-galactopyranuronate-(1-4)-methyl alpha-D-galactopyranuronate-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Fries, M, Brocklehurst, K, Shevchik, V.E, Pickersgill, R.W. | | Deposit date: | 2006-11-07 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of the activity of the phytopathogen pectin methylesterase.
Embo J., 26, 2007
|
|
2NTQ
 
 | | Crystal structure of pectin methylesterase in complex with hexasaccharide VII | | Descriptor: | Pectinesterase A, methyl alpha-D-galactopyranuronate-(1-4)-methyl alpha-D-galactopyranuronate-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Fries, M, Brocklehurst, K, Shevchik, V.E, Pickersgill, R.W. | | Deposit date: | 2006-11-08 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis of the activity of the phytopathogen pectin methylesterase.
Embo J., 26, 2007
|
|
2NSP
 
 | | Crystal structure of pectin methylesterase D178A mutant in complex with hexasaccharide I | | Descriptor: | Pectinesterase A, methyl alpha-D-galactopyranuronate-(1-4)-methyl alpha-D-galactopyranuronate-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-methyl alpha-D-galactopyranuronate | | Authors: | Fries, M, Brocklehurst, K, Shevchik, V.E, Pickersgill, R.W. | | Deposit date: | 2006-11-06 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of the activity of the phytopathogen pectin methylesterase.
Embo J., 26, 2007
|
|
2NTP
 
 | | Crystal structure of pectin methylesterase in complex with hexasaccharide VI | | Descriptor: | Pectinesterase A, methyl alpha-D-galactopyranuronate-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid, methyl beta-D-galactopyranuronate-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Fries, M, Brocklehurst, K, Shevchik, V.E, Pickersgill, R.W. | | Deposit date: | 2006-11-08 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of the activity of the phytopathogen pectin methylesterase.
Embo J., 26, 2007
|
|
2NST
 
 | | Crystal structure of pectin methylesterase D178A mutant in complex with hexasaccharide II | | Descriptor: | Pectinesterase A, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-methyl alpha-D-galactopyranuronate-(1-4)-methyl alpha-D-galactopyranuronate-(1-4)-methyl alpha-D-galactopyranuronate-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Fries, M, Brocklehurst, K, Shevchik, V.E, Pickersgill, R.W. | | Deposit date: | 2006-11-06 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of the activity of the phytopathogen pectin methylesterase.
Embo J., 26, 2007
|
|
2NTB
 
 | | Crystal structure of pectin methylesterase in complex with hexasaccharide V | | Descriptor: | Pectinesterase A, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Fries, M, Brocklehurst, K, Shevchik, V.E, Pickersgill, R.W. | | Deposit date: | 2006-11-07 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis of the activity of the phytopathogen pectin methylesterase.
Embo J., 26, 2007
|
|
2NT6
 
 | | Crystal structure of pectin methylesterase D178A mutant in complex with hexasaccharide III | | Descriptor: | Pectinesterase A, methyl alpha-D-galactopyranuronate-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-methyl alpha-D-galactopyranuronate | | Authors: | Fries, M, Brocklehurst, K, Shevchik, V.E, Pickersgill, R.W. | | Deposit date: | 2006-11-07 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of the activity of the phytopathogen pectin methylesterase.
Embo J., 26, 2007
|
|
7SPZ
 
 | | Nucleotide-free Get3 in two open forms | | Descriptor: | ATPase ASNA1 homolog, ZINC ION | | Authors: | Fry, M.Y, Maggiolo, A.O, Clemons Jr, W.M. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structurally derived universal mechanism for the catalytic cycle of the tail-anchored targeting factor Get3.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SPY
 
 | | Get3 bound to ATP from G. intestinalis in the closed form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase ASNA1 homolog, MAGNESIUM ION, ... | | Authors: | Fry, M.Y, Maggiolo, A.O, Clemons Jr, W.M. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structurally derived universal mechanism for the catalytic cycle of the tail-anchored targeting factor Get3.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SQ0
 
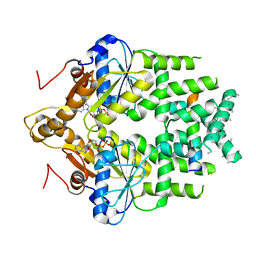 | | Get3 bound to ADP and the transmembrane domain of the tail-anchored protein Bos1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase ASNA1 homolog, MAGNESIUM ION, ... | | Authors: | Fry, M.Y, Maggiolo, A.O, Clemons Jr, W.M. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structurally derived universal mechanism for the catalytic cycle of the tail-anchored targeting factor Get3.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2WBJ
 
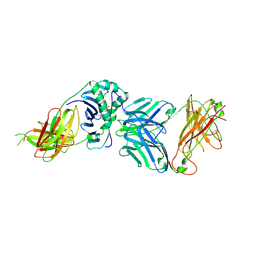 | | TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, DR ALPHA CHAIN, ... | | Authors: | Harkiolaki, M, Holmes, S.L, Svendsen, P, Gregersen, J.W, Jensen, L.T, McMahon, R, Friese, M.A, van Boxel, G, Etzensperger, R, Tzartos, J.S, Kranc, K, Sainsbury, S, Harlos, K, Mellins, E.D, Palace, J, Esiri, M.M, van der Merwe, P.A, Jones, E.Y, Fugger, L. | | Deposit date: | 2009-03-02 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | T Cell-Mediated Autoimmune Disease due to Low-Affinity Crossreactivity to Common Microbial Peptides.
Immunity, 30, 2009
|
|
3CBB
 
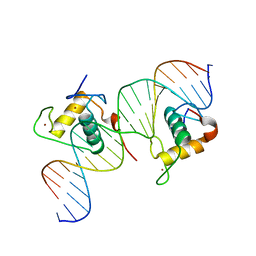 | | Crystal Structure of Hepatocyte Nuclear Factor 4alpha in complex with DNA: Diabetes Gene Product | | Descriptor: | Hepatocyte Nuclear Factor 4-alpha promoter element DNA, Hepatocyte Nuclear Factor 4-alpha, DNA binding domain, ... | | Authors: | Lu, P, Rha, G.B, Melikishvili, M, Adkins, B.C, Fried, M.G, Chi, Y.I. | | Deposit date: | 2008-02-21 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of natural promoter recognition by a unique nuclear receptor, HNF4alpha. Diabetes gene product.
J.Biol.Chem., 283, 2008
|
|
2QFT
 
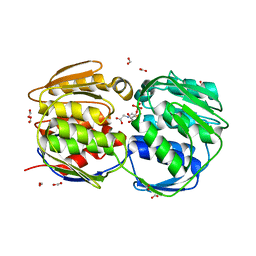 | |
2QFS
 
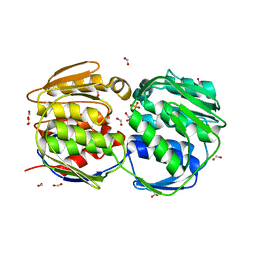 | | E.coli EPSP synthase Pro101Ser liganded with S3P | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, SHIKIMATE-3-PHOSPHATE | | Authors: | Schonbrunn, E, Healy-Fried, M.L. | | Deposit date: | 2007-06-28 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis of glyphosate tolerance resulting from mutations of Pro101 in Escherichia coli 5-enolpyruvylshikimate-3-phosphate synthase.
J.Biol.Chem., 282, 2007
|
|
2QFQ
 
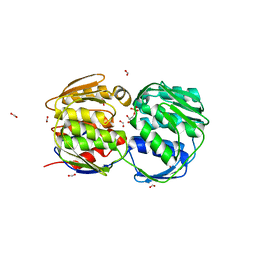 | | E. coli EPSP synthase Pro101Leu liganded with S3P | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, SHIKIMATE-3-PHOSPHATE | | Authors: | Schonbrunn, E, Healy-Fried, M.L. | | Deposit date: | 2007-06-27 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of glyphosate tolerance resulting from mutations of Pro101 in Escherichia coli 5-enolpyruvylshikimate-3-phosphate synthase.
J.Biol.Chem., 282, 2007
|
|
2QFU
 
 | |
2PQ9
 
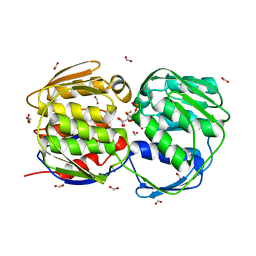 | | E. coli EPSPS liganded with (R)-difluoromethyl tetrahedral reaction intermediate analog | | Descriptor: | (3R,4S,5R)-5-[(1R)-1-CARBOXY-2,2-DIFLUORO-1-(PHOSPHONOOXY)ETHOXY]-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID | | Authors: | Healy-Fried, M.L, Funke, T, Han, H, Schonbrunn, E. | | Deposit date: | 2007-05-01 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Differential inhibition of class I and class II 5-enolpyruvylshikimate-3-phosphate synthases by tetrahedral reaction intermediate analogues.
Biochemistry, 46, 2007
|
|
2PQB
 
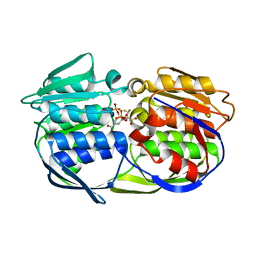 | | CP4 EPSPS liganded with (R)-difluoromethyl tetrahedral intermediate analog | | Descriptor: | (3R,4S,5R)-5-[(1R)-1-CARBOXY-2,2-DIFLUORO-1-(PHOSPHONOOXY)ETHOXY]-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Healy-Fried, M.L, Funke, T, Han, H, Schonbrunn, E. | | Deposit date: | 2007-05-01 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential inhibition of class I and class II 5-enolpyruvylshikimate-3-phosphate synthases by tetrahedral reaction intermediate analogues.
Biochemistry, 46, 2007
|
|
2PQC
 
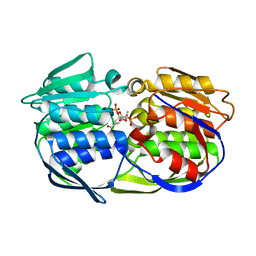 | | CP4 EPSPS liganded with (R)-phosphonate tetrahedral reaction intermediate analog | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, [3R-[3A,4A,5B(R*)]]-5-(1-CARBOXY-1-PHOSPHONOETHOXY)-4-HYDROXY-3-(PHOSPHONOOXY)-1-CYCLOHEXENE-1-CARBOXYLIC ACID | | Authors: | Healy-Fried, M.L, Funke, T, Han, H, Schonbrunn, E. | | Deposit date: | 2007-05-01 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Differential inhibition of class I and class II 5-enolpyruvylshikimate-3-phosphate synthases by tetrahedral reaction intermediate analogues.
Biochemistry, 46, 2007
|
|
2PQD
 
 | | A100G CP4 EPSPS liganded with (R)-difluoromethyl tetrahedral reaction intermediate analog | | Descriptor: | (3R,4S,5R)-5-[(1R)-1-CARBOXY-2,2-DIFLUORO-1-(PHOSPHONOOXY)ETHOXY]-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Healy-Fried, M.L, Funke, T, Han, H, Schonbrunn, E. | | Deposit date: | 2007-05-01 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Differential inhibition of class I and class II 5-enolpyruvylshikimate-3-phosphate synthases by tetrahedral reaction intermediate analogues.
Biochemistry, 46, 2007
|
|
2XPG
 
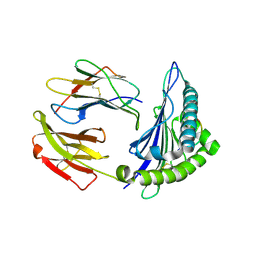 | | Crystal structure of a MHC class I-peptide complex | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, A-3 ALPHA CHAIN, ... | | Authors: | McMahon, R.M, Friis, L, Siebold, C, Friese, M.A, Fugger, L, Jones, E.Y. | | Deposit date: | 2010-08-26 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Hla-A0301 in Complex with a Peptide of Proteolipid Protein: Insights Into the Role of Hla-A Alleles in Susceptibility to Multiple Sclerosis
Acta Crystallogr.,Sect.D, 67, 2011
|
|
