1W85
 
 | | The crystal structure of pyruvate dehydrogenase E1 bound to the peripheral subunit binding domain of E2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE COMPONENT OF PYRUVATE, MAGNESIUM ION, ... | | Authors: | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A molecular switch and proton wire synchronize the active sites in thiamine enzymes.
Science, 306, 2004
|
|
1W88
 
 | | The crystal structure of pyruvate dehydrogenase E1(D180N,E183Q) bound to the peripheral subunit binding domain of E2 | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE COMPONENT OF PYRUVATE, MAGNESIUM ION, PYRUVATE DEHYDROGENASE E1 COMPONENT, ... | | Authors: | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Molecular Switch and Proton-Wire Synchronize the Active Sites in Thiamine-Dependent Enzymes
Science, 306, 2004
|
|
2JGD
 
 | | E. COLI 2-oxoglutarate dehydrogenase (E1o) | | Descriptor: | 2-OXOGLUTARATE DEHYDROGENASE E1 COMPONENT, ADENOSINE MONOPHOSPHATE | | Authors: | Frank, R.A.W, Price, A.J, Northrop, F.D, Perham, R.N, Luisi, B.F. | | Deposit date: | 2007-02-12 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the E1 Component of the Escherichia Coli 2-Oxoglutarate Dehydrogenase Multienzyme Complex.
J.Mol.Biol., 368, 2007
|
|
2BP7
 
 | | New crystal form of the Pseudomonas putida branched-chain dehydrogenase (E1) | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT | | Authors: | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | Deposit date: | 2005-04-18 | | Release date: | 2005-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Molecular Origins of Specificity in the Assembly of a Multienzyme Complex.
Structure, 13, 2005
|
|
6GBU
 
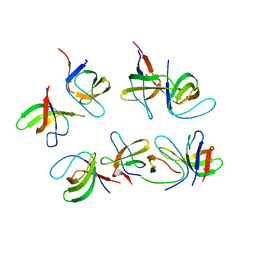 | | Crystal structure of the second SH3 domain of FCHSD2 (SH3-2) in complex with the fourth SH3 domain of ITSN1 (SH3d) | | Descriptor: | F-BAR and double SH3 domains protein 2, Intersectin-1 | | Authors: | Almeida-Souza, L, Frank, R, Garcia-Nafria, J, Colussi, A, Gunawardana, N, Johnson, C.M, Yu, M, Howard, G, Andrews, B, Vallis, Y, McMahon, H.T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | A Flat BAR Protein Promotes Actin Polymerization at the Base of Clathrin-Coated Pits.
Cell, 174, 2018
|
|
3DV0
 
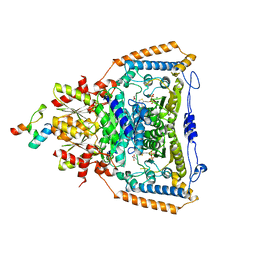 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-18 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
3DVA
 
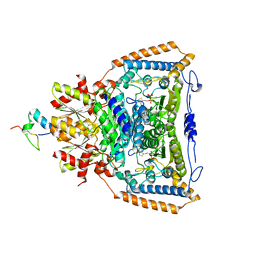 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-18 | | Release date: | 2009-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
3DUF
 
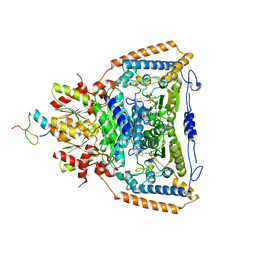 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-[(1R)-1-HYDROXYETHYL]-3-METHYL-2-THIENYL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-17 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
8BFB
 
 | | Sarkosyl-extracted AppNL-G-F Abeta42 fibril structure (Methoxy-X04-labelled mice) | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Wilkinson, M, Leistner, C, Burgess, A, Goodfellow, S, Deuchars, S, Ranson, N.A, Radford, S.E, Frank, R.A.W. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The in-tissue molecular architecture of beta-amyloid pathology in the mammalian brain.
Nat Commun, 14, 2023
|
|
8BFA
 
 | | Sarkosyl-extracted AppNL-G-F Abeta42 fibril structure | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Wilkinson, M, Leistner, C, Burgess, A, Goodfellow, S, Deuchars, S, Ranson, N.A, Radford, S.E, Frank, R.A.W. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The in-tissue molecular architecture of beta-amyloid pathology in the mammalian brain.
Nat Commun, 14, 2023
|
|
1ZTA
 
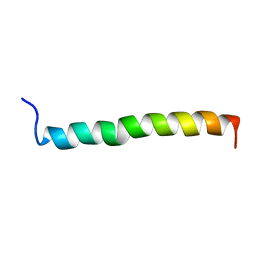 | | THE SOLUTION STRUCTURE OF A LEUCINE-ZIPPER MOTIF PEPTIDE | | Descriptor: | LEUCINE ZIPPER MONOMER | | Authors: | Saudek, V, Pastore, A, Castiglione Morelli, M.A, Frank, R, Gausepohl, H, Gibson, T. | | Deposit date: | 1990-10-11 | | Release date: | 1993-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a leucine-zipper motif peptide.
Protein Eng., 4, 1991
|
|
1ZTO
 
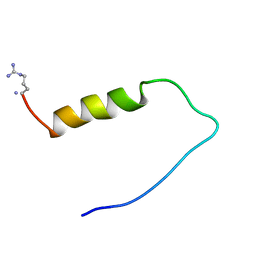 | | INACTIVATION GATE OF POTASSIUM CHANNEL RCK4, NMR, 8 STRUCTURES | | Descriptor: | POTASSIUM CHANNEL PROTEIN RCK4 | | Authors: | Antz, C, Geyer, M, Fakler, B, Schott, M, Frank, R, Guy, H.R, Ruppersberg, J.P, Kalbitzer, H.R. | | Deposit date: | 1996-11-15 | | Release date: | 1997-06-05 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure of inactivation gates from mammalian voltage-dependent potassium channels.
Nature, 385, 1997
|
|
1ZTN
 
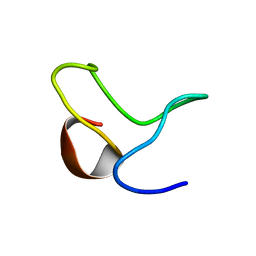 | | INACTIVATION GATE OF POTASSIUM CHANNEL RAW3, NMR, 8 STRUCTURES | | Descriptor: | Potassium voltage-gated channel subfamily C member 4 | | Authors: | Antz, C, Geyer, M, Fakler, B, Schott, M, Frank, R, Guy, H.R, Ruppersberg, J.P, Kalbitzer, H.R. | | Deposit date: | 1996-11-15 | | Release date: | 1997-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of inactivation gates from mammalian voltage-dependent potassium channels.
Nature, 385, 1997
|
|
1TIV
 
 | |
1B4I
 
 | | Control of K+ Channel Gating by protein phosphorylation: structural switches of the inactivation gate, NMR, 22 structures | | Descriptor: | POTASSIUM CHANNEL | | Authors: | Antz, C, Bauer, T, Kalbacher, H, Frank, R, Covarrubias, M, Kalbitzer, H.R, Ruppersberg, J.P, Baukrowitz, T, Fakler, B. | | Deposit date: | 1998-12-22 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Control of K+ channel gating by protein phosphorylation: structural switches of the inactivation gate.
Nat.Struct.Biol., 6, 1999
|
|
1B4G
 
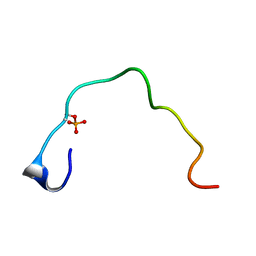 | | CONTROL OF K+ CHANNEL GATING BY PROTEIN PHOSPHORYLATION: STRUCTURAL SWITCHES OF THE INACTIVATION GATE, NMR, 22 STRUCTURES | | Descriptor: | POTASSIUM CHANNEL | | Authors: | Antz, C, Bauer, T, Kalbacher, H, Frank, R, Covarrubias, M, Kalbitzer, H.R, Ruppersberg, J.P, Baukrowitz, T, Fakler, B. | | Deposit date: | 1998-12-22 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Control of K+ channel gating by protein phosphorylation: structural switches of the inactivation gate.
Nat.Struct.Biol., 6, 1999
|
|
2WD1
 
 | | Human c-Met Kinase in complex with azaindole inhibitor | | Descriptor: | 1-[(2-NITROPHENYL)SULFONYL]-1H-PYRROLO[3,2-B]PYRIDINE-6-CARBOXAMIDE, GAMMA-BUTYROLACTONE, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | Porter, J, Lumb, S, Franklin, R.J, Gascon-Simorte, J.M, Calmiano, M, Le Riche, K, Lallemand, B, Keyaerts, J, Edwards, H, Maloney, A, Delgado, J, King, L, Foley, A, Lecomte, F, Reuberson, J, Meier, C, Batchelor, M. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-Azaindoles as Novel Inhibitors of C- met Kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
