1ETG
 
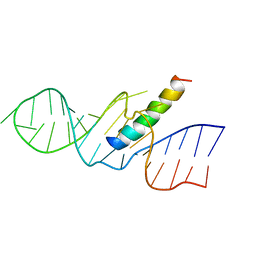 | | REV RESPONSE ELEMENT (RRE) RNA COMPLEXED WITH REV PEPTIDE, NMR, 19 STRUCTURES | | Descriptor: | REV PEPTIDE, REV RESPONSIVE ELEMENT RNA | | Authors: | Battiste, J.L, Mao, H, Rao, N.S, Tan, R, Muhandiram, D.R, Kay, L.E, Frankel, A.D, Willamson, J.R. | | Deposit date: | 1996-08-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex.
Science, 273, 1996
|
|
1ETF
 
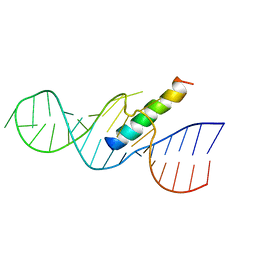 | | REV RESPONSE ELEMENT (RRE) RNA COMPLEXED WITH REV PEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | REV PEPTIDE, REV RESPONSE ELEMENT RNA | | Authors: | Battiste, J.L, Mao, H, Rao, N.S, Tan, R, Muhandiram, D.R, Kay, L.E, Frankel, A.D, Willamson, J.R. | | Deposit date: | 1996-08-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex.
Science, 273, 1996
|
|
2AH2
 
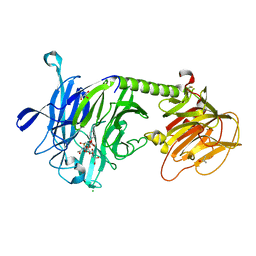 | | Trypanosoma cruzi trans-sialidase in complex with 2,3-difluorosialic acid (covalent intermediate) | | Descriptor: | 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Amaya, M.F, Watts, A.G, Damager, I, Wehenkel, A, Nguyen, T, Buschiazzo, A, Paris, G, Frasch, A.C, Withers, S.G, Alzari, P.M. | | Deposit date: | 2005-07-27 | | Release date: | 2005-08-23 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Trypanosoma cruzi trans-Sialidase
Structure, 12, 2004
|
|
8G2W
 
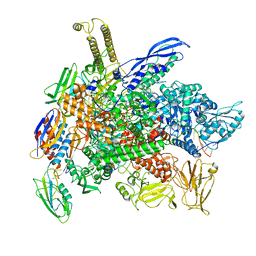 | | Cryo-EM structure of 3DVA component 2 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (31-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-02-06 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G4W
 
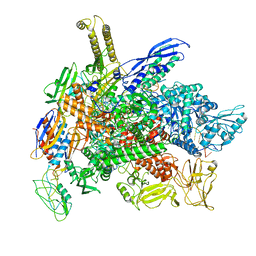 | | Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-mer), ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-02-10 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G8Z
 
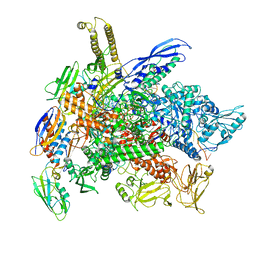 | | Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-MER), ... | | Authors: | Porta, J.C, Ohi, M.D, Walter, N.G, Frank, A.T, Deb, I, Meze, K. | | Deposit date: | 2023-02-20 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G1S
 
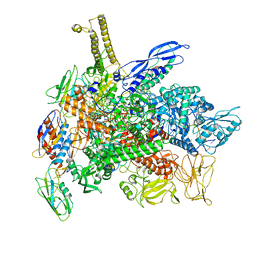 | | Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (31-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-02-02 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G00
 
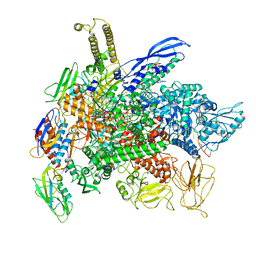 | | Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (31-MER), DNA (39-mer), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-01-31 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G7E
 
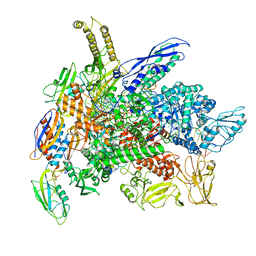 | | Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-mer), ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1OBT
 
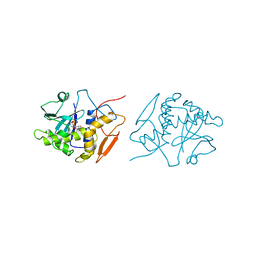 | | STRUCTURE OF RICIN A CHAIN MUTANT, COMPLEX WITH AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, RICIN A CHAIN | | Authors: | Day, P.J, Ernst, S.R, Frankel, A.E, Monzingo, A.F, Pascal, J.M, Svinth, M, Robertus, J.D. | | Deposit date: | 1996-06-22 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and activity of an active site substitution of ricin A chain.
Biochemistry, 35, 1996
|
|
6EEB
 
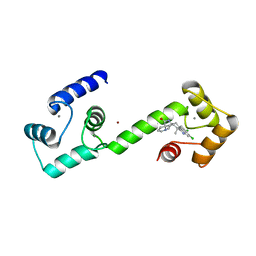 | | Calmodulin in complex with malbrancheamide | | Descriptor: | (5aS,12aS,13aS)-8,9-dichloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CALCIUM ION, Calmodulin-1, ... | | Authors: | Beyett, T.S, Fraley, A.E, Tesmer, J.J.G. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Perturbation of the interactions of calmodulin with GRK5 using a natural product chemical probe.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1OBS
 
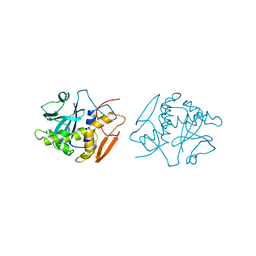 | | STRUCTURE OF RICIN A CHAIN MUTANT | | Descriptor: | RICIN A CHAIN | | Authors: | Day, P.J, Ernst, S.R, Frankel, A.E, Monzingo, A.F, Pascal, J.M, Svinth, M, Robertus, J.D. | | Deposit date: | 1996-06-25 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and activity of an active site substitution of ricin A chain.
Biochemistry, 35, 1996
|
|
4CSD
 
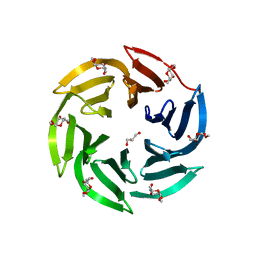 | | Structure of Monomeric Ralstonia solanacearum lectin | | Descriptor: | FUCOSE-BINDING LECTIN PROTEIN, GLYCEROL, methyl alpha-L-fucopyranoside | | Authors: | Arnaud, J, Trundle, K, Claudinon, J, Audfray, A, Varrot, A, Romer, W, Imberty, A. | | Deposit date: | 2014-03-06 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Membrane Deformation by Neolectins with Engineered Glycolipid Binding Sites.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1ZBN
 
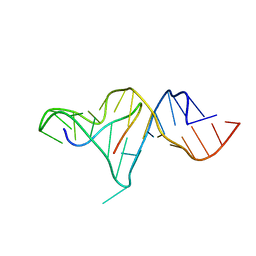 | |
7U4A
 
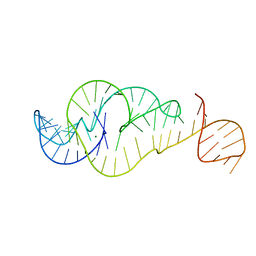 | | Crystal Structure of Zika virus xrRNA1 mutant | | Descriptor: | MAGNESIUM ION, RNA (70-MER) | | Authors: | Thompson, R.D, Carbaugh, D.L, Nielsen, J.R, Witt, C, Meganck, R.M, Rangadurai, A, Zhao, B, Bonin, J.P, Nathan, N.T, Marzluff, W.F, Frank, A.T, Lazear, H.M, Zhang, Q. | | Deposit date: | 2022-02-28 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Dynamic Basis of Xrn1 Resistance in Mosquito-borne Flavivirus RNA
To Be Published
|
|
6QDW
 
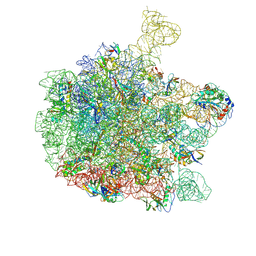 | | Cryo-EM structure of the 50S ribosomal subunit at 2.83 Angstroms with modeled GBC SecM peptide | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Schulte, L, Reitz, J, Hodirnau, V.V, Kudlinzki, D, Mao, J, Glaubitz, C, Frangakis, A, Schwalbe, H. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cysteine oxidation and disulfide formation in the ribosomal exit tunnel.
Nat Commun, 11, 2020
|
|
5M3F
 
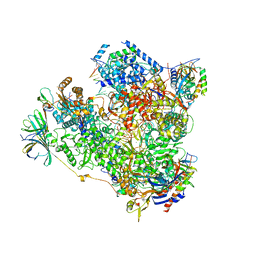 | | Yeast RNA polymerase I elongation complex at 3.8A | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Neyer, S, Kunz, M, Geiss, C, Hantsche, M, Hodirnau, V.-V, Seybert, A, Engel, C, Scheffer, M.P, Cramer, P, Frangakis, A.S. | | Deposit date: | 2016-10-14 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of RNA polymerase I transcribing ribosomal DNA genes.
Nature, 540, 2016
|
|
4BNA
 
 | |
1P7N
 
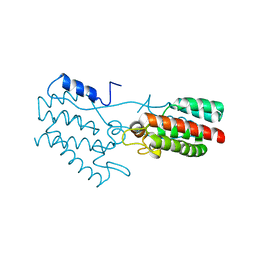 | | Dimeric Rous Sarcoma virus Capsid protein structure with an upstream 25-amino acid residue extension of C-terminal of Gag p10 protein | | Descriptor: | GAG POLYPROTEIN CAPSID PROTEIN P27 | | Authors: | Nandhagopal, N, Simpson, A.A, Johnson, M.C, Francisco, A.B, Schatz, G.W, Rossmann, M.G, Vogt, V.M. | | Deposit date: | 2003-05-02 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dimeric rous sarcoma virus capsid protein structure relevant to immature gag assembly
J.Mol.Biol., 335, 2004
|
|
1ROC
 
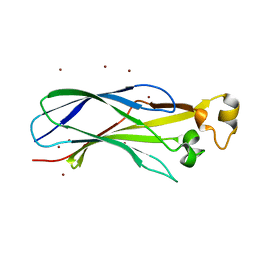 | | Crystal structure of the histone deposition protein Asf1 | | Descriptor: | Anti-silencing protein 1, BROMIDE ION | | Authors: | Daganzo, S.M, Erzberger, J.P, Lam, W.M, Skordalakes, E, Zhang, R, Franco, A.A, Brill, S.J, Adams, P.D, Berger, J.M, Kaufman, P.D. | | Deposit date: | 2003-12-02 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the conserved core of histone deposition protein Asf1.
Curr.Biol., 13, 2003
|
|
4LUZ
 
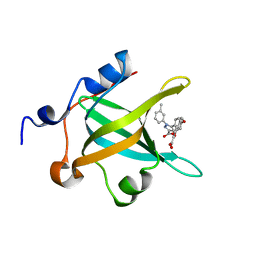 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-(4-{[4-(5-carboxyfuran-2-yl)benzyl]oxy}phenyl)-1-(3-methylphenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
4LWC
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-[3-chloro-4-({4-[1-(3,4-dichlorophenyl)-1H-pyrazol-5-yl]benzyl}carbamothioyl)phenyl]furan-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-26 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
1I9F
 
 | | STRUCTURAL CHARACTERIZATION OF THE COMPLEX OF THE REV RESPONSE ELEMENT RNA WITH A SELECTED PEPTIDE | | Descriptor: | REV RESPONSE ELEMENT RNA, RSG-1.2 PEPTIDE | | Authors: | Zhang, Q, Harada, K, Cho, H.S, Frankel, A, Wemmer, D.E. | | Deposit date: | 2001-03-19 | | Release date: | 2001-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the complex of the Rev response element RNA with a selected peptide.
Chem.Biol., 8, 2001
|
|
1S0I
 
 | | Trypanosoma cruzi trans-sialidase in complex with sialyl-lactose (Michaelis complex) | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, trans-sialidase | | Authors: | Amaya, M.F, Watts, A.G, Damager, I, Wehenkel, A, Nguyen, T, Buschiazzo, A, Paris, G, Frasch, A.C, Withers, S.G, Alzari, P.M. | | Deposit date: | 2003-12-31 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Trypanosoma cruzi trans-Sialidase.
Structure, 12, 2004
|
|
4O0A
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-{4-[({[4-(5-carboxyfuran-2-yl)-2-chlorophenyl]carbonothioyl}amino)methyl]phenyl}-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejniczak, E.T, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-12-13 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J. Med. Chem., 56, 2013
|
|
