4S3C
 
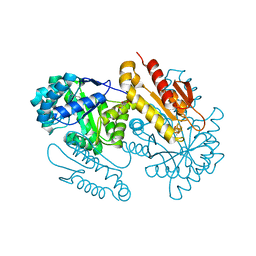 | | IspG in complex with Epoxide Intermediate | | Descriptor: | (3S)-1,3-dihydroxy-4-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}-2-methylbut-2-ylium, carbokation intermediate, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
4S3E
 
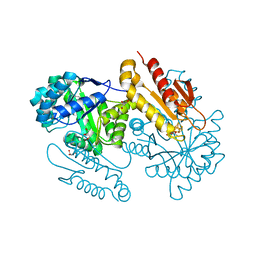 | | IspG in complex with Inhibitor 7 (compound 1061) | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
4S3B
 
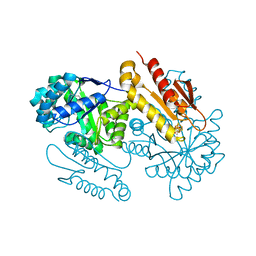 | | IspG in complex with Intermediate II | | Descriptor: | (3S)-1,3-dihydroxy-4-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}-2-methylbut-2-ylium, carbokation intermediate, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
6LU1
 
 | | Cyanobacterial PSI Monomer from T. elongatus by Single Particle CRYO-EM at 3.2 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kurisu, G, Coruh, O, Tanaka, H, Gerle, C, Kawamoto, A, Kato, T, Namba, K, Nowaczyk, M.M, Rogner, M, Misumi, Y, Frank, A, Eithar, E.M. | | Deposit date: | 2020-01-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus reveals red chlorophyll cluster.
Commun Biol, 4, 2021
|
|
4S3D
 
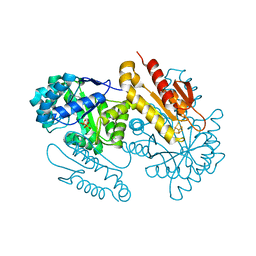 | | IspG in complex with PPi | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, DIPHOSPHATE, FE3-S4 CLUSTER | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
4S3F
 
 | | IspG in complex with Inhibitor 8 (compound 1077) | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, FE3-S4 CLUSTER, but-3-yn-1-yl trihydrogen diphosphate | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
4S3A
 
 | | IspG in complex with Intermediate I | | Descriptor: | (3S)-1,3-dihydroxy-4-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}-2-methylbut-2-ylium, carbokation intermediate, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
7U4A
 
 | | Crystal Structure of Zika virus xrRNA1 mutant | | Descriptor: | MAGNESIUM ION, RNA (70-MER) | | Authors: | Thompson, R.D, Carbaugh, D.L, Nielsen, J.R, Witt, C, Meganck, R.M, Rangadurai, A, Zhao, B, Bonin, J.P, Nathan, N.T, Marzluff, W.F, Frank, A.T, Lazear, H.M, Zhang, Q. | | Deposit date: | 2022-02-28 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Dynamic Basis of Xrn1 Resistance in Mosquito-borne Flavivirus RNA
To Be Published
|
|
8P1I
 
 | | Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 2.97 Angstrom resolution | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL), 3-chloranyl-2,6-di(piperazin-4-ium-1-yl)quinoline, Efflux pump membrane transporter, ... | | Authors: | Boernsen, C, Mueller, R.T, Pos, K.M, Frangakis, A.S. | | Deposit date: | 2023-05-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Pyridylpiperazine efflux pump inhibitor boosts in vivo antibiotic efficacy against K. pneumoniae.
Embo Mol Med, 16, 2024
|
|
7B8Q
 
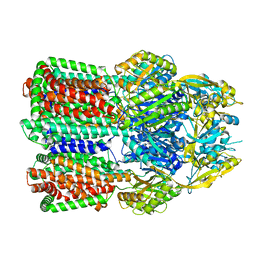 | | Acinetobacter baumannii multidrug transporter AdeB in L*OO state | | Descriptor: | Efflux pump membrane transporter | | Authors: | Ornik-Cha, A, Reitz, J, Seybert, A, Frangakis, A, Pos, K.M. | | Deposit date: | 2020-12-13 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms.
Nat Commun, 12, 2021
|
|
7B8P
 
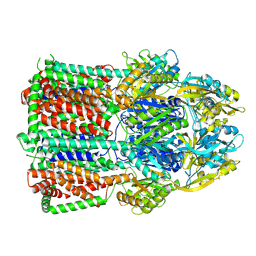 | | Acinetobacter baumannii multidrug transporter AdeB in OOO state | | Descriptor: | Efflux pump membrane transporter | | Authors: | Ornik-Cha, A, Reitz, J, Seybert, A, Frangakis, A, Pos, K.M. | | Deposit date: | 2020-12-13 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms.
Nat Commun, 12, 2021
|
|
8P4I
 
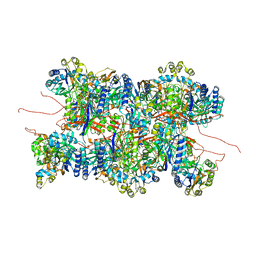 | | Cyanide dihydratase from Bacillus pumilus C1 | | Descriptor: | Cyanide dihydratase | | Authors: | Mulelu, A.E, Reitz, J, van Rooyen, J.M, Scheffer, M, Frangakis, A.S, Dlamini, L.S, Woodward, J.D, Benedik, M.J, Sewell, B.T. | | Deposit date: | 2023-05-22 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | The Role of Histidine Residues in the Oligomerization of Cyanide Dihydratase from Bacillus pumilus C1
To Be Published
|
|
8UDT
 
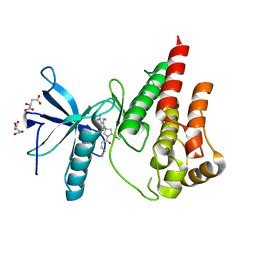 | | The X-RAY co-crystal structure of human FGFR3 and KIN-3248 | | Descriptor: | 3-[(1-cyclopropyl-4,6-difluoro-1H-benzimidazol-5-yl)ethynyl]-1-[(3R,5R)-5-(methoxymethyl)-1-propanoylpyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, D-MALATE, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.829 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
8PBZ
 
 | |
8UDV
 
 | | The X-RAY co-crystal structure of human FGFR3 V555M and Compound 17 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, Fibroblast growth factor receptor 3, ... | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
8UDU
 
 | | The X-RAY co-crystal structure of human FGFR3 and Compound 17 | | Descriptor: | 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, CHLORIDE ION, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
7S01
 
 | | X-ray structure of the phage AR9 non-virion RNA polymerase holoenzyme in complex with a forked oligonucleotide containing the P077 promoter | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase beta subunit, DNA-directed RNA polymerase beta' subunit, ... | | Authors: | Leiman, P.G, Sokolova, M.L, Gordeeva, J, Fraser, A, Severinov, K.V. | | Deposit date: | 2021-08-28 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of template strand deoxyuridine promoter recognition by a viral RNA polymerase.
Nat Commun, 13, 2022
|
|
7S00
 
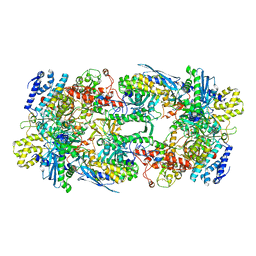 | | X-ray structure of the phage AR9 non-virion RNA polymerase core | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase beta subunit, DNA-directed RNA polymerase beta' subunit, ... | | Authors: | Leiman, P.G, Sokolova, M.L, Fraser, A. | | Deposit date: | 2021-08-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of template strand deoxyuridine promoter recognition by a viral RNA polymerase.
Nat Commun, 13, 2022
|
|
7A7D
 
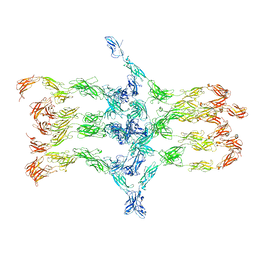 | | Cadherin fit into cryo-ET map | | Descriptor: | Desmocollin-2, Desmoglein-2 | | Authors: | Sikora, M, Ermel, U.H, Seybold, A, Kunz, M, Calloni, G, Reitz, J, Vabulas, R.M, Hummer, G, Frangakis, A.S. | | Deposit date: | 2020-08-28 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Desmosome architecture derived from molecular dynamics simulations and cryo-electron tomography.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7UM0
 
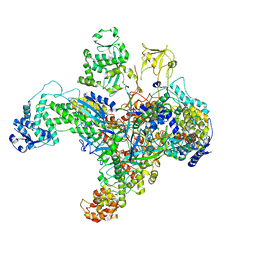 | | Structure of the phage AR9 non-virion RNA polymerase holoenzyme in complex with two DNA oligonucleotides containing the AR9 P077 promoter as determined by cryo-EM | | Descriptor: | DNA (5'-D(P*GP*UP*U)-3'), DNA-directed RNA polymerase, DNA-directed RNA polymerase beta subunit, ... | | Authors: | Leiman, P.G, Fraser, A, Sokolova, M.L. | | Deposit date: | 2022-04-05 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of template strand deoxyuridine promoter recognition by a viral RNA polymerase.
Nat Commun, 13, 2022
|
|
8PC0
 
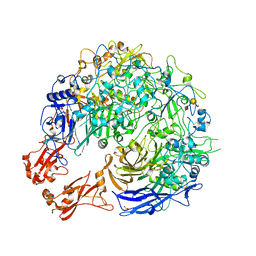 | | Sub-tomogram average of the open conformation of the Nap adhesion complex from the human pathogen Mycoplasma genitalium. | | Descriptor: | Adhesin P1, Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-1,5-anhydro-D-glucitol, ... | | Authors: | Sprankel, L, Scheffer, M.P, Frangakis, A.F. | | Deposit date: | 2023-06-09 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Cryo-electron tomography reveals the binding and release states of the major adhesion complex from Mycoplasma genitalium.
Plos Pathog., 19, 2023
|
|
7UM1
 
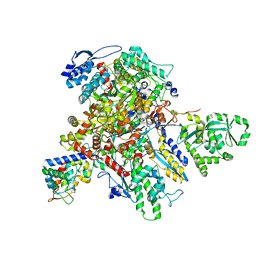 | | Structure of bacteriophage AR9 non-virion RNAP polymerase holoenzyme determined by cryo-EM | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase beta subunit, DNA-directed RNA polymerase beta' subunit, ... | | Authors: | Leiman, P.G, Fraser, A, Sokolova, M.L. | | Deposit date: | 2022-04-05 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of template strand deoxyuridine promoter recognition by a viral RNA polymerase.
Nat Commun, 13, 2022
|
|
5M3F
 
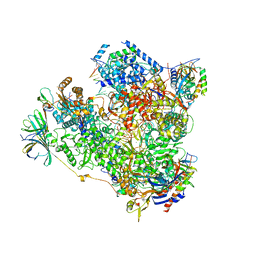 | | Yeast RNA polymerase I elongation complex at 3.8A | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Neyer, S, Kunz, M, Geiss, C, Hantsche, M, Hodirnau, V.-V, Seybert, A, Engel, C, Scheffer, M.P, Cramer, P, Frangakis, A.S. | | Deposit date: | 2016-10-14 | | Release date: | 2016-11-23 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of RNA polymerase I transcribing ribosomal DNA genes.
Nature, 540, 2016
|
|
5M3M
 
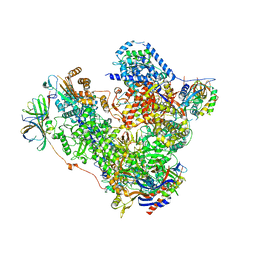 | | Free monomeric RNA polymerase I at 4.0A resolution | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Neyer, S, Kunz, M, Geiss, C, Hantsche, M, Hodirnau, V.-V, Seybert, A, Engel, C, Scheffer, M.P, Cramer, P, Frangakis, A.S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of RNA polymerase I transcribing ribosomal DNA genes.
Nature, 540, 2016
|
|
1ZBN
 
 | |
