1TF3
 
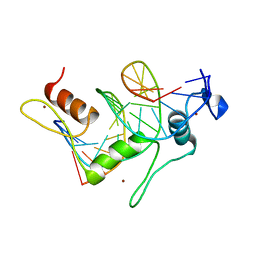 | | TFIIIA FINGER 1-3 BOUND TO DNA, NMR, 22 STRUCTURES | | Descriptor: | 5S RNA GENE, TRANSCRIPTION FACTOR IIIA, ZINC ION | | Authors: | Foster, M.P, Wuttke, D.S, Radhakrishnan, I, Case, D.A, Gottesfeld, J.M, Wright, P.E. | | Deposit date: | 1997-07-01 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Domain packing and dynamics in the DNA complex of the N-terminal zinc fingers of TFIIIA.
Nat.Struct.Biol., 4, 1997
|
|
5VXB
 
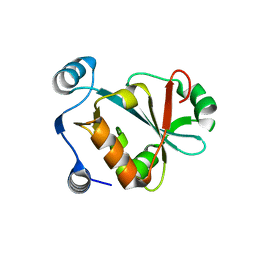 | |
9BU5
 
 | | CC ProXp-ala apo solution structure | | Descriptor: | DNA-binding protein, putative | | Authors: | Duran, A.D, Foster, M.P. | | Deposit date: | 2024-05-16 | | Release date: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR-based solution structure of the Caulobacter crescentus ProXp-ala trans-editing enzyme.
Biomol.Nmr Assign., 2024
|
|
2AV5
 
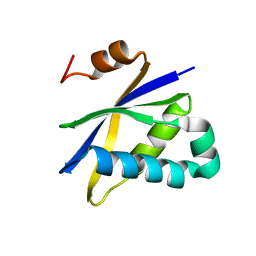 | | Crystal structure of Pyrococcus furiosus Pop5, an archaeal Ribonuclease P protein | | Descriptor: | Ribonuclease P protein component 2 | | Authors: | Wilson, R.C, Bohlen, C.J, Foster, M.P, Bell, C.E. | | Deposit date: | 2005-08-29 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of Pfu Pop5, an archaeal RNase P protein.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7RHY
 
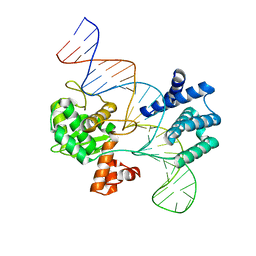 | |
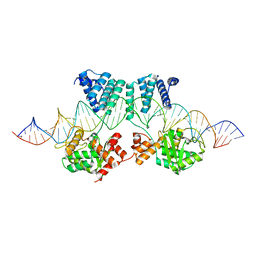
7RHZ
 
 | |
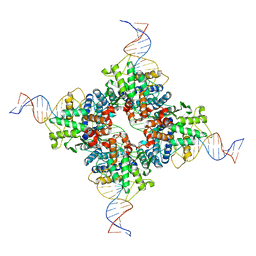
7RHX
 
 | |
1OQK
 
 | | Structure of Mth11: A homologue of human RNase P protein Rpp29 | | Descriptor: | conserved protein MTH11 | | Authors: | Boomershine, W.P, McElroy, C.A, Tsai, H, Gopalan, V, Foster, M.P. | | Deposit date: | 2003-03-10 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Mth11/Mth Rpp29, an essential protein subunit of archaeal and eukaryotic RNase P.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1AS5
 
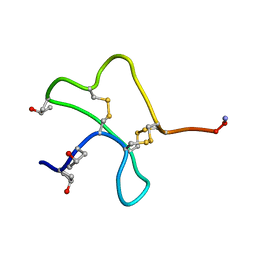 | | SOLUTION STRUCTURE OF CONOTOXIN Y-PIIIE FROM CONUS PURPURASCENS, NMR, 14 STRUCTURES | | Descriptor: | CONOTOXIN Y-PIIIE | | Authors: | Mitchell, S.S, Shon, K, Foster, M.P, Olivera, B.M, Ireland, C.M. | | Deposit date: | 1997-08-13 | | Release date: | 1998-10-14 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of conotoxin psi-PIIIE, an acetylcholine gated ion channel antagonist.
Biochemistry, 37, 1998
|
|
1RXR
 
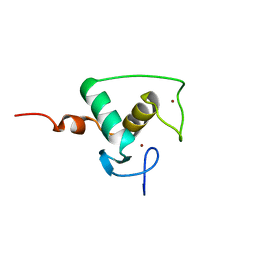 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE RETINOID X RECEPTOR DNA BINDING DOMAIN, NMR, 20 STRUCTURE | | Descriptor: | RETINOIC ACID RECEPTOR-ALPHA, ZINC ION | | Authors: | Holmbeck, S.M.A, Foster, M.P, Casimiro, D.R, Sem, D.S, Dyson, H.J, Wright, P.E. | | Deposit date: | 1998-06-12 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the retinoid X receptor DNA-binding domain.
J.Mol.Biol., 281, 1998
|
|
2KMN
 
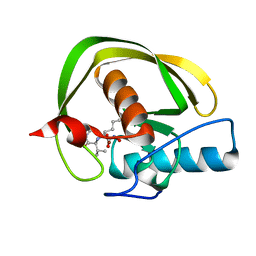 | |
2KI7
 
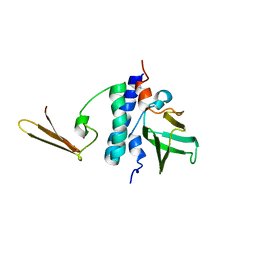 | | The solution structure of RPP29-RPP21 complex from Pyrococcus furiosus | | Descriptor: | Ribonuclease P protein component 1, Ribonuclease P protein component 4, ZINC ION | | Authors: | Xu, Y, Foster, M.P. | | Deposit date: | 2009-04-28 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an archaeal RNase P binary protein complex: formation of the 30-kDa complex between Pyrococcus furiosus RPP21 and RPP29 is accompanied by coupled protein folding and highlights critical features for protein-protein and protein-RNA interactions.
J.Mol.Biol., 393, 2009
|
|
2KO8
 
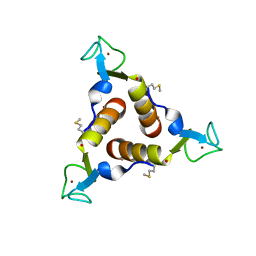 | | The Structure of Anti-TRAP | | Descriptor: | Tryptophan RNA-binding attenuator protein inhibitory protein, ZINC ION | | Authors: | McElroy, C.A, Gollnick, P, Foster, M.P. | | Deposit date: | 2009-09-12 | | Release date: | 2010-09-22 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the B. subtilis Anti-TRAP trimer and its Interaction with TRAP
To be Published
|
|
2M16
 
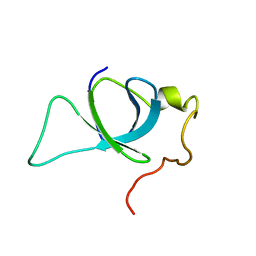 | | P75/LEDGF PWWP Domain | | Descriptor: | PC4 and SFRS1-interacting protein | | Authors: | Crowe, B.L, Foster, M.P. | | Deposit date: | 2012-11-18 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes.
Nucleic Acids Res., 41, 2013
|
|
2N3K
 
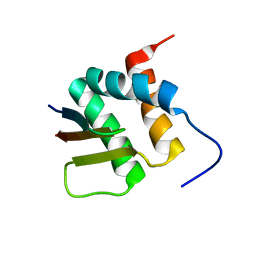 | | Human Brd4 ET domain in complex with MLV Integrase C-term | | Descriptor: | Bromodomain-containing protein 4, MLV integrase | | Authors: | Crowe, B.L, Foster, M.P. | | Deposit date: | 2015-06-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Brd4 ET domain bound to a C-terminal motif from gamma-retroviral integrases reveals a conserved mechanism of interaction.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2OXO
 
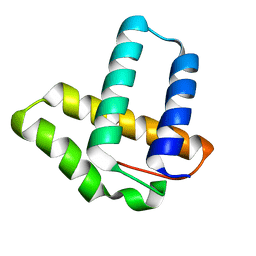 | |
2K3R
 
 | | Pfu Rpp21 structure and assignments | | Descriptor: | Ribonuclease P protein component 4, ZINC ION | | Authors: | Amero, C.D, Foster, M.P, Boomershine, W.P, Xu, Y. | | Deposit date: | 2008-05-15 | | Release date: | 2008-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pyrococcus furiosus RPP21, a component of the archaeal RNase P holoenzyme, and interactions with its RPP29 protein partner.
Biochemistry, 47, 2008
|
|
