1EFZ
 
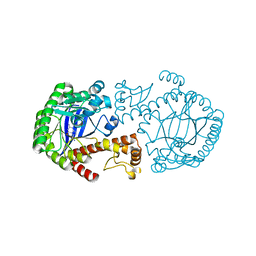 | | MUTAGENESIS AND CRYSTALLOGRAPHIC STUDIES OF ZYMOMONAS MOBILIS TRNA-GUANINE TRANSGLYCOSYLASE TO ELUCIDATE THE ROLE OF SERINE 103 FOR ENZYMATIC ACTIVITY | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Gradler, U, Ficner, R, Garcia, G.A, Stubbs, M.T, Klebe, G, Reuter, K. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase to elucidate the role of serine 103 for enzymatic activity.
FEBS Lett., 454, 1999
|
|
1QGV
 
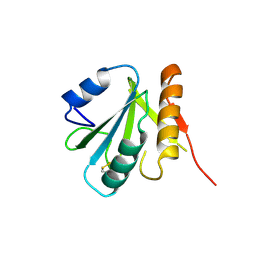 | | HUMAN SPLICEOSOMAL PROTEIN U5-15KD | | Descriptor: | SPLICEOSOMAL PROTEIN U5-15KD | | Authors: | Reuter, K, Nottrott, S, Fabrizio, P, Luehrmann, R, Ficner, R. | | Deposit date: | 1999-05-06 | | Release date: | 1999-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification, characterization and crystal structure analysis of the human spliceosomal U5 snRNP-specific 15 kD protein.
J.Mol.Biol., 294, 1999
|
|
7NQ4
 
 | |
1QR0
 
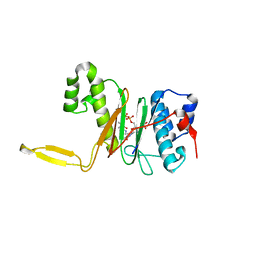 | | CRYSTAL STRUCTURE OF THE 4'-PHOSPHOPANTETHEINYL TRANSFERASE SFP-COENZYME A COMPLEX | | Descriptor: | 4'-PHOSPHOPANTETHEINYL TRANSFERASE SFP, COENZYME A, MAGNESIUM ION | | Authors: | Reuter, K, Mofid, R.M, Marahiel, A.M, Ficner, R. | | Deposit date: | 1999-06-17 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the surfactin synthetase-activating enzyme sfp: a prototype of the 4'-phosphopantetheinyl transferase superfamily.
EMBO J., 18, 1999
|
|
9G0G
 
 | | BsCdaA in complex with Compound 7 | | Descriptor: | CHLORIDE ION, Cyclic di-AMP synthase CdaA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2024-07-08 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic fragment screen of the c-di-AMP-synthesizing enzyme CdaA from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
3NC0
 
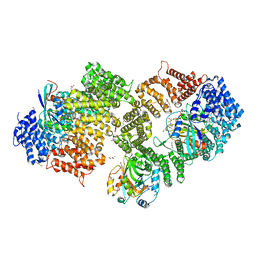 | | Crystal structure of the HIV-1 Rev NES-CRM1-RanGTP nuclear export complex (crystal II) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Exportin-1, GLYCEROL, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
9F79
 
 | | Crystal structure of the S. cerevisiae eIF2beta N-terminal tail bound to the C-terminal domain of eIF5 | | Descriptor: | Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 5 | | Authors: | Kuhle, B, Marintchev, A, Ficner, R. | | Deposit date: | 2024-05-03 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the interactions of eIF2 beta with eIF5, eIF2B, and 5MP1 and their regulation by CK2.
Rna, 31, 2025
|
|
2QNA
 
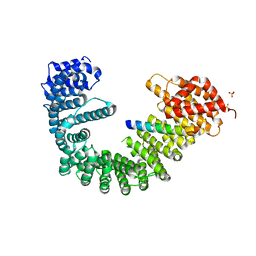 | | Crystal structure of human Importin-beta (127-876) in complex with the IBB-domain of Snurportin1 (1-65) | | Descriptor: | Importin subunit beta-1, SULFATE ION, Snurportin-1 | | Authors: | Wohlwend, D, Strasser, A, Dickmanns, A, Ficner, R. | | Deposit date: | 2007-07-18 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural basis for RanGTP independent entry of spliceosomal U snRNPs into the nucleus.
J.Mol.Biol., 374, 2007
|
|
1E7K
 
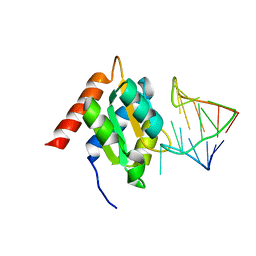 | | Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment | | Descriptor: | 15.5 KD RNA BINDING PROTEIN, RNA (5'-R(*GP*CP*CP*AP*AP*UP*GP*AP*GP*GP*UP*UP*UP* AP*UP*CP*CP*GP*AP*GP*G*C(-3') | | Authors: | Vidovic, I, Nottrott, S, Harthmuth, K, Luhrmann, R, Ficner, R. | | Deposit date: | 2000-08-29 | | Release date: | 2001-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Spliceosomal 15.5Kd Protein Bound to a U4 Snrna Fragment
Mol.Cell, 6, 2000
|
|
2VCG
 
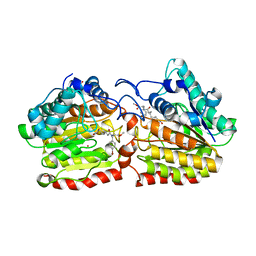 | | Crystal structure of a HDAC-like protein HDAH from Bordetella sp. with the bound inhibitor ST-17 | | Descriptor: | CHLORIDE ION, GLYCEROL, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Dickmanns, A, Strasser, A, Ficner, R. | | Deposit date: | 2007-09-24 | | Release date: | 2008-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phenylalanine-Containing Hydroxamic Acids as Selective Inhibitors of Class Iib Histone Deacetylases (Hdacs).
Bioorg.Med.Chem., 16, 2008
|
|
6I3P
 
 | |
2GH6
 
 | | Crystal structure of a HDAC-like protein with 9,9,9-trifluoro-8-oxo-N-phenylnonan amide bound | | Descriptor: | 9,9,9-TRIFLUORO-8-OXO-N-PHENYLNONANAMIDE, Histone deacetylase-like amidohydrolase, POTASSIUM ION, ... | | Authors: | Nielsen, T.K, Hildmann, C, Riester, D, Wegener, D, Schwienhorst, A, Ficner, R. | | Deposit date: | 2006-03-26 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Complex structure of a bacterial class 2 histone deacetylase homologue with a trifluoromethylketone inhibitor.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1PUD
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-06-28 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of tRNA-guanine transglycosylase: RNA modification by base exchange.
EMBO J., 15, 1996
|
|
3JU4
 
 | | Crystal Structure Analysis of EndosialidaseNF at 0.98 A Resolution | | Descriptor: | CHLORIDE ION, Endo-N-acetylneuraminidase, N-acetyl-beta-neuraminic acid, ... | | Authors: | Schulz, E.C, Neuman, P, Gerardy-Schahn, R, Sheldrick, G.M, Ficner, R. | | Deposit date: | 2009-09-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structure analysis of endosialidase NF at 0.98 A resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6ZM2
 
 | |
5DTU
 
 | |
7NZJ
 
 | | Structure of bsTrmB apo | | Descriptor: | GLYCEROL, SODIUM ION, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Blersch, K.F, Ficner, R, Neumann, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural model of the M7G46 Methyltransferase TrmB in complex with tRNA.
Rna Biol., 18, 2021
|
|
7NZI
 
 | | TrmB complex with SAH | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Blersch, K.F, Ficner, R, Neumann, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-15 | | Last modified: | 2025-12-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural model of the M7G46 Methyltransferase TrmB in complex with tRNA.
Rna Biol., 18, 2021
|
|
7NYB
 
 | | TrmB complex with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Blersch, K.F, Ficner, R, Neumann, P. | | Deposit date: | 2021-03-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural model of the M7G46 Methyltransferase TrmB in complex with tRNA.
Rna Biol., 18, 2021
|
|
1WKD
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
1WKE
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
7B51
 
 | | Crystal structure of human CRM1 covalently modified by 2-mercaptoethanol at Cys528 | | Descriptor: | BETA-MERCAPTOETHANOL, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Shaikhqasem, A, Ficner, R. | | Deposit date: | 2020-12-03 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of human CRM1, covalently modified by 2-mercaptoethanol on Cys528, in complex with RanGTP.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
1WKF
 
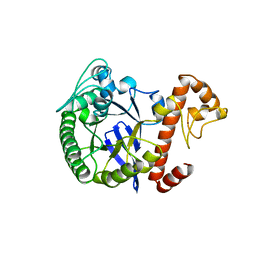 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
5EK8
 
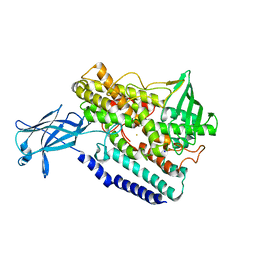 | | Crystal structure of a 9R-lipoxygenase from Cyanothece PCC8801 at 2.7 Angstroms | | Descriptor: | FE (II) ION, Lipoxygenase, SODIUM ION | | Authors: | Feussner, I, Ficner, R, Neumann, P, Newie, J, Andreou, A, Einsle, O. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a lipoxygenase from Cyanothece sp. may reveal novel features for substrate acquisition.
J.Lipid Res., 57, 2016
|
|
3CTR
 
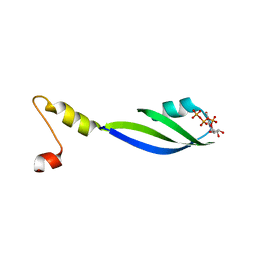 | | Crystal structure of the RRM-domain of the poly(A)-specific ribonuclease PARN bound to m7GTP | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Poly(A)-specific ribonuclease PARN | | Authors: | Monecke, T, Schell, S, Dickmanns, A, Ficner, R. | | Deposit date: | 2008-04-14 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the RRM domain of poly(A)-specific ribonuclease reveals a novel m(7)G-cap-binding mode.
J.Mol.Biol., 382, 2008
|
|
