3HJU
 
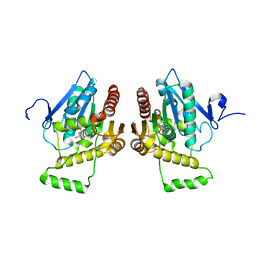 | | Crystal structure of human monoglyceride lipase | | Descriptor: | GLYCEROL, Monoglyceride lipase | | Authors: | Labar, G, Bauvois, C, Borel, F, Ferrer, J.-L, Wouters, J, Lambert, D.M. | | Deposit date: | 2009-05-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Human Monoacylglycerol Lipase, a Key Actor in Endocannabinoid Signaling
Chembiochem, 11, 2010
|
|
1KNK
 
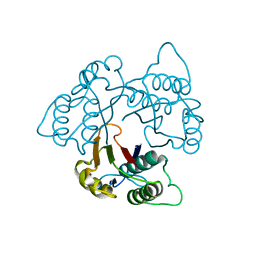 | | Crystal Structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate Synthase (ispF) from E. coli involved in Mevalonate-Independent Isoprenoid Biosynthesis | | Descriptor: | 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, MANGANESE (II) ION | | Authors: | Richard, S.B, Ferrer, J.L, Bowman, M.E, Lillo, A.M, Tetzlaff, C.N, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-12-18 | | Release date: | 2002-06-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase. An enzyme in the mevalonate-independent isoprenoid biosynthetic pathway.
J.Biol.Chem., 277, 2002
|
|
1ZC2
 
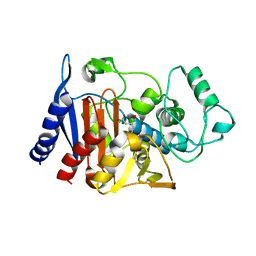 | | Crystal Structure of plasmid-encoded class C beta-lactamase CMY-2 complexed with citrate molecule | | Descriptor: | CITRIC ACID, beta-lactamase class C | | Authors: | Bauvois, C, Jacquamet, L, Fieulaine, S, Frere, J.-M, Galleni, M, Ferrer, J.-L. | | Deposit date: | 2005-04-10 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystallographic structure of plasmid-encoded CMY-2 beta-lactamase revealed citrate molecule in the active site.
To be Published
|
|
3F8N
 
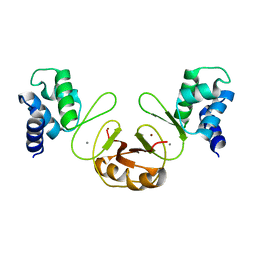 | | Crystal structure of PerR-Zn-Mn | | Descriptor: | MANGANESE (II) ION, Peroxide operon regulator, ZINC ION | | Authors: | Traore, D.A.K, Ferrer, J.-L, Jacquamet, L, Duarte, V, Latour, J.-M. | | Deposit date: | 2008-11-13 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding
Mol.Microbiol., 73, 2009
|
|
1KYZ
 
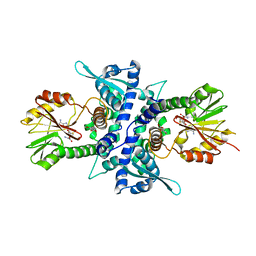 | | Crystal Structure Analysis of Caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase Ferulic Acid Complex | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Caffeic acid 3-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Kota, P, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2002-02-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase.
Plant Cell, 14, 2002
|
|
5A4D
 
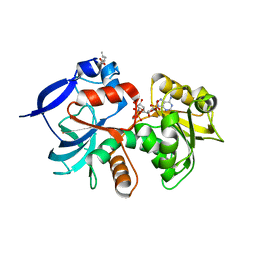 | | Crystal structure of the chloroplastic gamma-ketol reductase from Arabidopsis thaliana bound to 13KOTE and NADP | | Descriptor: | (13-oxo-9(Z),11(E),15(Z)-octadecatrienoic acid), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PUTATIVE QUINONE-OXIDOREDUCTASE HOMOLOG, ... | | Authors: | Mas-y-mas, S, Curien, G, Giustini, C, Rolland, N, Ferrer, J.L, Cobessi, D. | | Deposit date: | 2015-06-08 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Crystal Structure of the Chloroplastic Oxoene Reductase ceQORH from Arabidopsis thaliana.
Front Plant Sci, 8, 2017
|
|
3L76
 
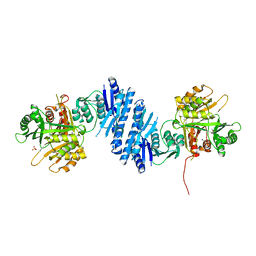 | | Crystal Structure of Aspartate Kinase from Synechocystis | | Descriptor: | Aspartokinase, LYSINE, SULFATE ION, ... | | Authors: | Robin, A, Cobessi, D, Curien, G, Robert-Genthon, M, Ferrer, J.-L, Dumas, R. | | Deposit date: | 2009-12-28 | | Release date: | 2010-06-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | A new mode of dimerization of allosteric enzymes with ACT domains revealed by the crystal structure of the aspartate kinase from Cyanobacteria
J.Mol.Biol., 399, 2010
|
|
3CIA
 
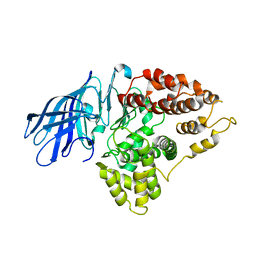 | | Crystal structure of cold-aminopeptidase from Colwellia psychrerythraea | | Descriptor: | ZINC ION, cold-active aminopeptidase | | Authors: | Bauvois, C, Jacquamet, L, Borel, F, Ferrer, J.-L. | | Deposit date: | 2008-03-11 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Cold-active Aminopeptidase from Colwellia psychrerythraea, a Close Structural Homologue of the Human Bifunctional Leukotriene A4 Hydrolase.
J.Biol.Chem., 283, 2008
|
|
2P0U
 
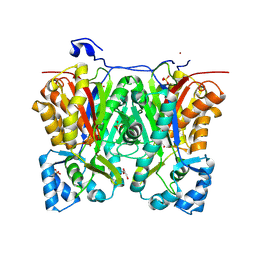 | | crystal structure of Marchantia polymorpha stilbenecarboxylate synthase 2 (STCS2) | | Descriptor: | GLYCEROL, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Borel, F, Schroeder, G, Schroeder, J, Ferrer, J.-L. | | Deposit date: | 2007-03-01 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | crystal structure of Marchantia polymorpha stilbenecarboxylate synthase 2 (STCS2)
To be Published
|
|
1KYW
 
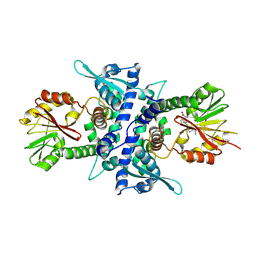 | | Crystal Structure Analysis of Caffeic Acid/5-hydroxyferulic acid 3/5-O-methyltransferase in complex with 5-hydroxyconiferaldehyde | | Descriptor: | 5-(3,3-DIHYDROXYPROPENY)-3-METHOXY-BENZENE-1,2-DIOL, Caffeic acid 3-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Kota, P, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2002-02-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase.
Plant Cell, 14, 2002
|
|
4BVA
 
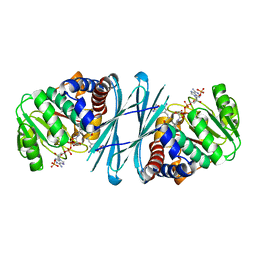 | | Crystal structure of the NADPH-T3 form of mouse Mu-crystallin. | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Borel, F, Hachi, I, Palencia, A, Gaillard, M.C, Ferrer, J.L. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Mouse Mu-Crystallin Complexed with Nadph and the T3 Thyroid Hormone
FEBS J., 281, 2014
|
|
2FA0
 
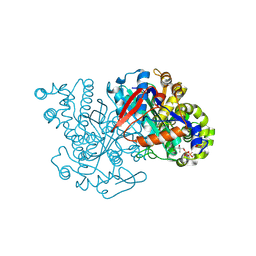 | | HMG-CoA synthase from Brassica juncea in complex with HMG-CoA and covalently bound to HMG-CoA | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, HMG-CoA synthase | | Authors: | Pojer, F, Ferrer, J.L, Richard, S.B, Noel, J.P. | | Deposit date: | 2005-12-06 | | Release date: | 2006-07-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2F9A
 
 | | HMG-CoA synthase from Brassica juncea in complex with F-244 | | Descriptor: | (7R,12R,13R)-13-formyl-12,14-dihydroxy-3,5,7-trimethyltetradeca-2,4-dienoic acid, 3-Hydroxy-3-methylglutaryl coenzyme A synthase 1 | | Authors: | Pojer, F, Ferrer, J.L, Richard, S.B, Noel, J.P. | | Deposit date: | 2005-12-05 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FA3
 
 | | HMG-CoA synthase from Brassica juncea in complex with acetyl-CoA and acetyl-cys117. | | Descriptor: | ACETYL COENZYME *A, HMG-CoA synthase | | Authors: | Pojer, F, Ferrer, J.L, Richard, S.B, Noel, J.P. | | Deposit date: | 2005-12-06 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3DOX
 
 | | X-ray structure of HIV-1 protease in situ product complex | | Descriptor: | A PEPTIDE SUBSTRATE-PIV, A PEPTIDE SUBSTRATE-SQNY, HIV-1 PROTEASE | | Authors: | Hosur, M.V, Ferrer, J.-L, Das, A, Prashar, V, Bihani, S. | | Deposit date: | 2008-07-07 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of HIV-1 protease in situ product complex
Proteins, 74, 2009
|
|
2F82
 
 | |
3ZGN
 
 | | Crystal structures of Escherichia coli IspH in complex with TMBPP a potent inhibitor of the methylerythritol phosphate pathway | | Descriptor: | (2E)-3-methyl-4-sulfanylbut-2-en-1-yl trihydrogen diphosphate, 4-HYDROXY-3-METHYLBUT-2-ENYL DIPHOSPHATE REDUCTASE, IRON/SULFUR CLUSTER | | Authors: | Borel, F, Barbier, E, Kratsutsky, S, Janthawornpong, K, Rohmer, M, Dale Poulter, C, Ferrer, J.L, Seemann, M. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Further Insight into Crystal Structures of Escherichia coli IspH/LytB in Complex with Two Potent Inhibitors of the MEP Pathway: A Starting Point for Rational Design of New Antimicrobials.
Chembiochem, 18, 2017
|
|
3ZGL
 
 | | Crystal structures of Escherichia coli IspH in complex with AMBPP a potent inhibitor of the methylerythritol phosphate pathway | | Descriptor: | (2E)-4-amino-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-HYDROXY-3-METHYLBUT-2-ENYL DIPHOSPHATE REDUCTASE, IRON/SULFUR CLUSTER | | Authors: | Borel, F, Barbier, E, Kratsutsky, S, Janthawornpong, K, Rohmer, M, Dale Poulter, C, Ferrer, J.L, Seemann, M. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Further Insight into Crystal Structures of Escherichia coli IspH/LytB in Complex with Two Potent Inhibitors of the MEP Pathway: A Starting Point for Rational Design of New Antimicrobials.
Chembiochem, 18, 2017
|
|
4BV9
 
 | | Crystal structure of the NADPH form of mouse Mu-crystallin. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Borel, F, Hachi, I, Palencia, A, Gaillard, M.C, Ferrer, J.L. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Crystal Structure of Mouse Mu-Crystallin Complexed with Nadph and the T3 Thyroid Hormone
FEBS J., 281, 2014
|
|
5A3V
 
 | | Crystal structure of the chloroplastic gamma-ketol reductase from Arabidopsis thaliana | | Descriptor: | PUTATIVE QUINONE-OXIDOREDUCTASE HOMOLOG, CHLOROPLASTIC | | Authors: | Mas-y-mas, S, Curien, G, Giustini, C, Rolland, N, Ferrer, J.L, Cobessi, D. | | Deposit date: | 2015-06-03 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of the Chloroplastic Oxoene Reductase ceQORH from Arabidopsis thaliana.
Front Plant Sci, 8, 2017
|
|
2F1K
 
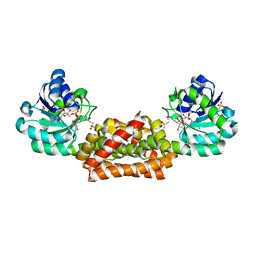 | | Crystal structure of Synechocystis arogenate dehydrogenase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, prephenate dehydrogenase | | Authors: | Legrand, P, Dumas, R, Seux, M, Rippert, P, Ravelli, R, Ferrer, J.-L, Matringe, M. | | Deposit date: | 2005-11-14 | | Release date: | 2006-05-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biochemical Characterization and Crystal Structure of Synechocystis Arogenate Dehydrogenase Provide Insights into Catalytic Reaction
Structure, 14, 2006
|
|
4BV8
 
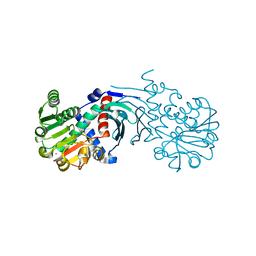 | | Crystal structure of the apo form of mouse Mu-crystallin. | | Descriptor: | GLYCEROL, POTASSIUM ION, THIOMORPHOLINE-CARBOXYLATE DEHYDROGENASE | | Authors: | Borel, F, Hachi, I, Palencia, A, Gaillard, M.C, Ferrer, J.L. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Mouse Mu-Crystallin Complexed with Nadph and the T3 Thyroid Hormone
FEBS J., 281, 2014
|
|
4A0G
 
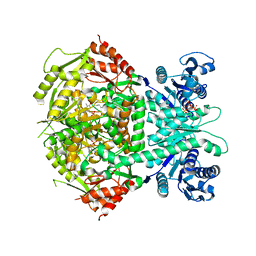 | | Structure of bifunctional DAPA aminotransferase-DTB synthetase from Arabidopsis thaliana in its apo form. | | Descriptor: | ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
4A0F
 
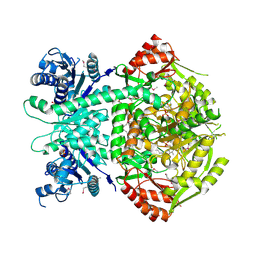 | | Structure of selenomethionine substituted bifunctional DAPA aminotransferase-dethiobiotin synthetase from Arabidopsis thaliana in its apo form. | | Descriptor: | ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (2.714 Å) | | Cite: | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
4A0H
 
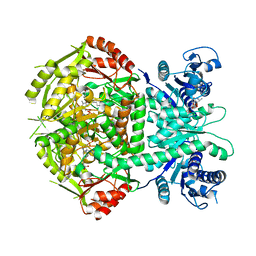 | | Structure of bifunctional DAPA aminotransferase-DTB synthetase from Arabidopsis thaliana bound to 7-keto 8-amino pelargonic acid (KAPA) | | Descriptor: | 7-KETO-8-AMINOPELARGONIC ACID, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, L(+)-TARTARIC ACID, ... | | Authors: | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
