6L87
 
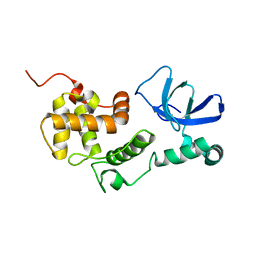 | |
1RKN
 
 | | Solution structure of 1-110 fragment of Staphylococcal Nuclease with G88W mutation | | Descriptor: | Thermonuclease | | Authors: | Liu, D.S, Feng, Y.G, Ye, K.Q, Shan, L, Wang, J.F. | | Deposit date: | 2003-11-22 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
1YYB
 
 | |
8H97
 
 | | GH86 agarase Aga86A_Wa | | Descriptor: | Beta-agarase, CALCIUM ION, HEXAETHYLENE GLYCOL | | Authors: | Zhang, Y.Y, Dong, S, Feng, Y.G, Chang, Y.G. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural characterization on a beta-agarase Aga86A_Wa from Wenyingzhuangia aestuarii reveals the prevalent methyl-galactose accommodation capacity of GH86 enzymes at subsite -1.
Carbohydr Polym, 306, 2023
|
|
5YJ6
 
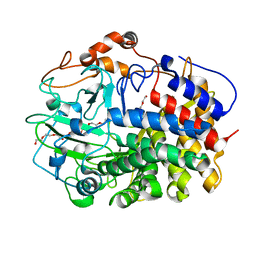 | | The exoglucanase CelS from Clostridium thermocellum | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36-dodecaoxaoctatriacontane-1,38-diol, Dockerin type I repeat-containing protein | | Authors: | Liu, Y.J, Liu, S.Y, Dong, S, Li, R.M, Feng, Y.G, Cui, Q. | | Deposit date: | 2017-10-09 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Determination of the native features of the exoglucanase Cel48S from Clostridium thermocellum
Biotechnol Biofuels, 11, 2018
|
|
5YJ7
 
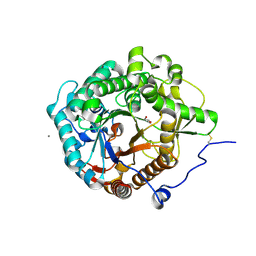 | | Structural insight into the beta-GH1 glucosidase BGLN1 from oleaginous microalgae Nannochloropsis | | Descriptor: | CALCIUM ION, GLYCEROL, Glycoside hydrolase | | Authors: | Dong, S, Liu, Y.J, Zhou, H.X, Xiao, Y, Xu, J, Cui, Q, Wang, X.Q, Feng, Y.G. | | Deposit date: | 2017-10-09 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insight into a GH1 beta-glucosidase from the oleaginous microalga, Nannochloropsis oceanica.
Int.J.Biol.Macromol., 170, 2021
|
|
8HDJ
 
 | |
5ZQI
 
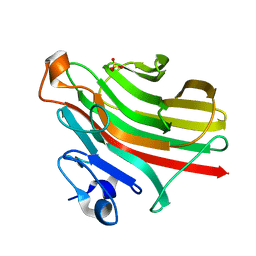 | | Alginate lyase AlgAT5 from Polysaccharide Lyase family 7 | | Descriptor: | Alginate lyase AlgAT5, SULFATE ION | | Authors: | Su, H, Dong, S, Feng, Y.G, Ji, S.Q, Lu, M, Li, F.L. | | Deposit date: | 2018-04-19 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Alginate lyase AlgAT5 from Polysaccharide Lyase family 7
To Be Published
|
|
8WBS
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases KlCESH[L]-D48N complexed with sulfate ions | | Descriptor: | (S)-2-haloacid dehalogenase, CALCIUM ION, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBO
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] mutant D18N complexed with sulfate ions | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBT
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases KlCESH[L] mutant D48N complexed with L-TA | | Descriptor: | (S)-2-haloacid dehalogenase, CALCIUM ION, GLYCEROL, ... | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBR
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases KlCESH[L] | | Descriptor: | (S)-2-haloacid dehalogenase, CALCIUM ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBK
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBQ
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] mutant E212Q complexed with L-TA. | | Descriptor: | Epoxide hydrolase, L(+)-TARTARIC ACID | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBP
 
 | |
8WBN
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] mutant D193N | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBL
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] complexed with sulfate ions | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBM
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] mutant D193A complexed with sulfate ions | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
7CCF
 
 | | Mechanism insights on steroselective oxidation of phosphorylated ethylphenols with cytochrome P450 CreJ | | Descriptor: | (3-ethylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Du, L, Li, S.Y, Feng, Y.G. | | Deposit date: | 2020-06-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Selective Oxidation of Phosphorylated Ethylphenols by Cytochrome P450 Monooxygenase CreJ.
Appl.Environ.Microbiol., 87, 2021
|
|
8XTF
 
 | | Crystal structure of methyltransferase MpaG' in complex with SAH and FDHMP-3C | | Descriptor: | 4-farnesyl-3,5-dihydroxy-6-methylphthalide-3C, O-methyltransferase mpaG', S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | You, C, Pan, Y.J, Li, S.Y, Feng, Y.G. | | Deposit date: | 2024-01-10 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis for substrate flexibility of the O-methyltransferase MpaG' involved in mycophenolic acid biosynthesis.
Protein Sci., 33, 2024
|
|
8XTG
 
 | | Crystal structure of methyltransferase MpaG' in complex with SAH and DMMPA | | Descriptor: | O-desmethyl mycophenolic acid, O-methyltransferase mpaG', S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | You, C, Pan, Y.J, Li, S.Y, Feng, Y.G. | | Deposit date: | 2024-01-10 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for substrate flexibility of the O-methyltransferase MpaG' involved in mycophenolic acid biosynthesis.
Protein Sci., 33, 2024
|
|
8XTE
 
 | | Crystal structure of methyltransferase MpaG' in complex with SAH and FDHMP | | Descriptor: | 4-farnesyl-3,5-dihydroxy-6-methylphthalide, O-methyltransferase mpaG', S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | You, C, Pan, Y.J, Li, S.Y, Feng, Y.G. | | Deposit date: | 2024-01-10 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for substrate flexibility of the O-methyltransferase MpaG' involved in mycophenolic acid biosynthesis.
Protein Sci., 33, 2024
|
|
7DMK
 
 | | PL6 alginate lyase BcAlyPL6 | | Descriptor: | BcAlyPL6, CALCIUM ION, MALONATE ION | | Authors: | Dong, S, Wang, B, Ma, X.Q, Li, F.L, Feng, Y.G. | | Deposit date: | 2020-12-04 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | Structural basis for the exolytic activity of polysaccharide lyase family 6 alginate lyase BcAlyPL6 from human gut microbe Bacteroides clarus.
Biochem.Biophys.Res.Commun., 547, 2021
|
|
