1HOV
 
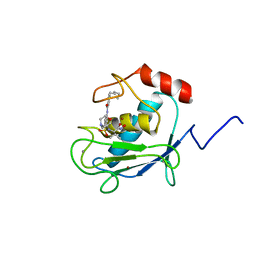 | | SOLUTION STRUCTURE OF A CATALYTIC DOMAIN OF MMP-2 COMPLEXED WITH SC-74020 | | Descriptor: | CALCIUM ION, MATRIX METALLOPROTEINASE-2, N-{4-[(1-HYDROXYCARBAMOYL-2-METHYL-PROPYL)-(2-MORPHOLIN-4-YL-ETHYL)-SULFAMOYL]-4-PENTYL-BENZAMIDE, ... | | Authors: | Feng, Y, Likos, J.J, Zhu, L, Woodward, H, Munie, G, McDonald, J.J, Stevens, A.M, Howard, C.P, De Crescenzo, G.A, Welsch, D, Shieh, H.-S, Stallings, W.C. | | Deposit date: | 2000-12-11 | | Release date: | 2001-12-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the catalytic domain of matrix metalloproteinase-2 complexed with a hydroxamic acid inhibitor
Biochim.Biophys.Acta, 1598, 2002
|
|
4MEY
 
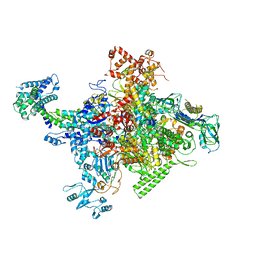 | | Crystal structure of Escherichia coli RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Feng, Y, Zhang, Y, Arnold, E, Ebright, R.H. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.948 Å) | | Cite: | Transcription inhibition by the depsipeptide antibiotic salinamide A.
Elife, 3, 2014
|
|
4R5Y
 
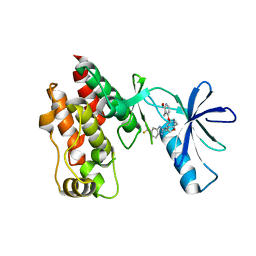 | | The complex structure of Braf V600E kinase domain with a novel Braf inhibitor | | Descriptor: | 5-({(1R,1aS,6bR)-1-[5-(trifluoromethyl)-1H-benzimidazol-2-yl]-1a,6b-dihydro-1H-cyclopropa[b][1]benzofuran-5-yl}oxy)-3,4-dihydro-1,8-naphthyridin-2(1H)-one, Serine/threonine-protein kinase B-raf | | Authors: | Feng, Y, Peng, H, Zhang, Y, Liu, Y, Wei, M. | | Deposit date: | 2014-08-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | BGB-283, a Novel RAF Kinase and EGFR Inhibitor, Displays Potent Antitumor Activity in BRAF-Mutated Colorectal Cancers.
Mol.Cancer Ther., 14, 2015
|
|
6LP2
 
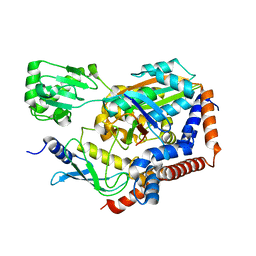 | | Structure of Lpg2148/UBE2N-Ub complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 N, Uncharacterized protein lpg2148 | | Authors: | Feng, Y, Wang, Y, Huang, Y, Li, D. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.479 Å) | | Cite: | Structure of Lpg2148/UBE2N-Ub complex
To Be Published
|
|
6SXN
 
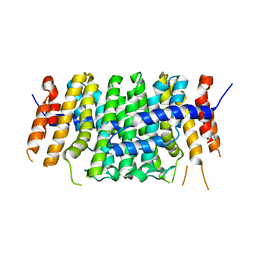 | | Crystal structure of P212121 apo form of CrtE | | Descriptor: | Geranylgeranyl pyrophosphate synthase | | Authors: | Feng, Y, Morgan, R.M.L, Nixon, P.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase (CrtE) Involved in Cyanobacterial Terpenoid Biosynthesis.
Front Plant Sci, 11, 2020
|
|
1JLI
 
 | |
6SXL
 
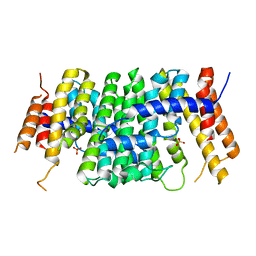 | | Crystal structure of CrtE | | Descriptor: | Geranylgeranyl pyrophosphate synthase, PHOSPHATE ION | | Authors: | Feng, Y, Morgan, R.M.L, Nixon, P.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase (CrtE) Involved in Cyanobacterial Terpenoid Biosynthesis.
Front Plant Sci, 11, 2020
|
|
7XSP
 
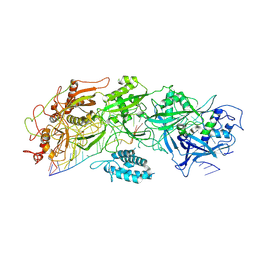 | | Structure of gRAMP-target RNA | | Descriptor: | RAMP superfamily protein, RNA (35-MER), RNA (5'-R(P*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*G)-3'), ... | | Authors: | Feng, Y, Zhang, L.X. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XSR
 
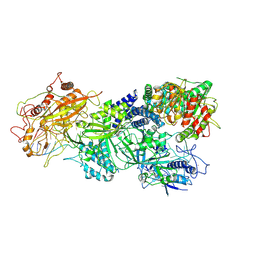 | | Structure of Craspase-target RNA | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zhang, L. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XSQ
 
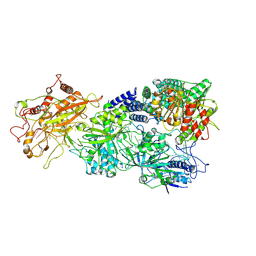 | | Structure of the Craspase | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zhang, L. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XSS
 
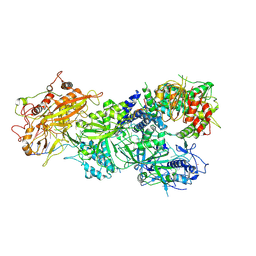 | | Structure of Craspase-CTR | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zang, L.X. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
6IGM
 
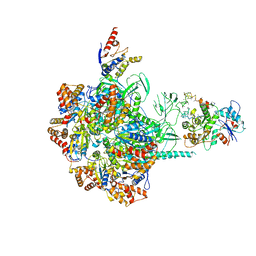 | | Cryo-EM Structure of Human SRCAP Complex | | Descriptor: | Actin-related protein 6, Helicase SRCAP, RuvB-like 1, ... | | Authors: | Feng, Y, Tian, Y, Wu, Z, Xu, Y. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of human SRCAP complex.
Cell Res., 28, 2018
|
|
4ZH3
 
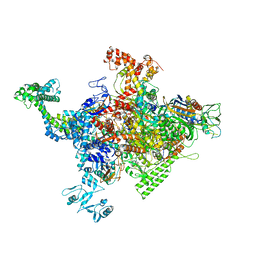 | |
4ZH4
 
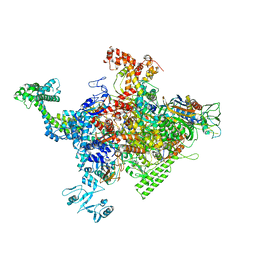 | | Crystal structure of Escherichia coli RNA polymerase in complex with CBRP18 | | Descriptor: | 5-(4-fluorophenyl)-4-[4-fluoro-3-(trifluoromethyl)phenyl]-1H-pyrazole, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Feng, Y, Ebright, R.H. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.993 Å) | | Cite: | Structural Basis of Transcription Inhibition by CBR Hydroxamidines and CBR Pyrazoles.
Structure, 23, 2015
|
|
4ZH2
 
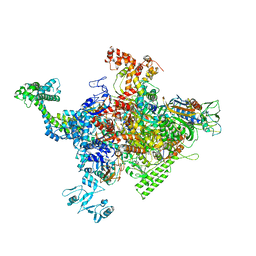 | |
2MK5
 
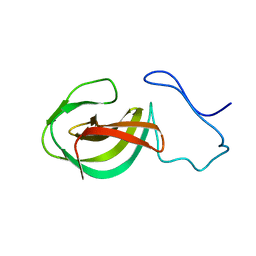 | | Solution structure of a protein domain | | Descriptor: | Endolysin | | Authors: | Feng, Y, Gu, J. | | Deposit date: | 2014-01-24 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and biochemical characterization reveals LysGH15 as an unprecedented "EF-hand-like" calcium-binding phage lysin.
Plos Pathog., 10, 2014
|
|
9IK2
 
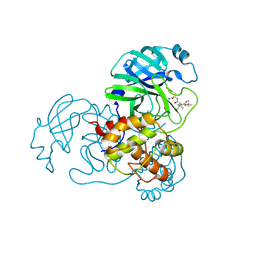 | | The co-crystal structure of SARS-CoV-2 Mpro in complex with compound H109 | | Descriptor: | 3C-like proteinase, tert-butyl N-[(2S)-1-[[(2S)-1-[[(2S)-1-azanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Feng, Y, Zheng, W.Y, Han, P, Fu, L.F, Qi, J.X. | | Deposit date: | 2024-06-26 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of a small molecule inhibitor of SARS-CoV-2 main protease with potent in vitro and in vivo antiviral activities
To Be Published
|
|
6LW4
 
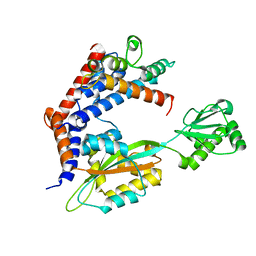 | |
6KGF
 
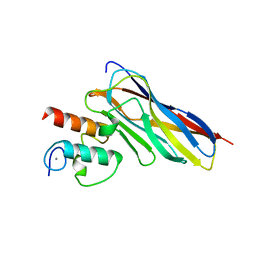 | | Crystal structure of CaDoc0917(R16D)-CaCohA2 complex at pH 8.2 | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION, Probably cellulosomal scaffolding protein, ... | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6KGC
 
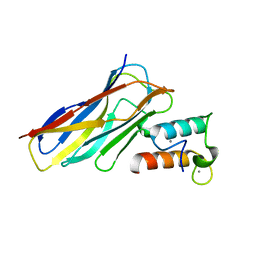 | | Crystal structure of CaDoc0917(R49D)-CaCohA2 complex at pH 5.4 | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION, Probably cellulosomal scaffolding protein, ... | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6INW
 
 | | A Pericyclic Reaction enzyme | | Descriptor: | O-methyltransferase lepI, S-ADENOSYLMETHIONINE | | Authors: | Feng, Y, Chang, M, Wang, H, Liu, Z, Zhou, Y. | | Deposit date: | 2018-10-28 | | Release date: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Crystal structure of the multifunctional SAM-dependent enzyme LepI provides insights into its catalytic mechanism.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
6KGD
 
 | | Crystal structure of CaDoc0917(R49D)-CaCohA2 complex at pH 8.0 | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION, Probably cellulosomal scaffolding protein, ... | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6KGE
 
 | | Crystal structure of CaDoc0917(R16D)-CaCohA2 complex at pH 5.5 | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION, Probably cellulosomal scaffolding protein, ... | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6KG8
 
 | | Solution structure of CaCohA2 from Clostridium acetobutylicum | | Descriptor: | Probably cellulosomal scaffolding protein, secreted cellulose-binding and cohesin domain | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6KG9
 
 | | Solution structure of CaDoc0917 from Clostridium acetobutylicum | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
