7OW8
 
 | | CryoEM structure of the ABC transporter BmrA E504A mutant in complex with ATP-Mg | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Gobet, A, Schoehn, G, Falson, P, Chaptal, V. | | Deposit date: | 2021-06-17 | | Release date: | 2022-01-19 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
9GSJ
 
 | | BmrA E504A in complex with Hoechst33342 | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Moissonnier, L, Zarkadas, E, Schoehn, G, Falson, P, Chaptal, V. | | Deposit date: | 2024-09-16 | | Release date: | 2025-03-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rhodamine6G and Hœchst33342 narrow BmrA conformational spectrum for a more efficient use of ATP.
Nat Commun, 16, 2025
|
|
7BG4
 
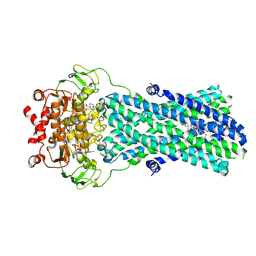 | | Multidrug resistance transporter BmrA mutant E504A bound with ATP, Mg, and Rhodamine 6G solved by Cryo-EM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding/permease protein BmrA, ... | | Authors: | Wiseman, B, Chaptal, V, Zampieri, V, Magnard, S, Hogbom, M, Falson, P. | | Deposit date: | 2021-01-05 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
6R72
 
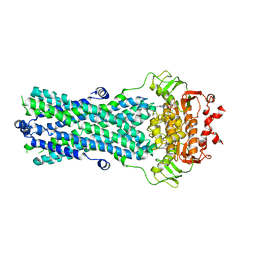 | | Crystal structure of BmrA-E504A in an outward-facing conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug exporter ATP-binding cassette | | Authors: | Chaptal, V, Zampieri, V, Kilburg, A, Magnard, S, Falson, P. | | Deposit date: | 2019-03-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
6R81
 
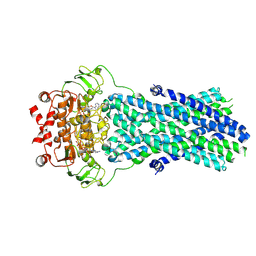 | | Multidrug resistance transporter BmrA mutant E504A bound with ATP and Mg solved by Cryo-EM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid A export ATP-binding/permease protein MsbA, MAGNESIUM ION | | Authors: | Wiseman, B, Chaptal, V, Zampieri, V, Magnard, S, Hogbom, M, Falson, P. | | Deposit date: | 2019-03-30 | | Release date: | 2020-05-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
4C3Z
 
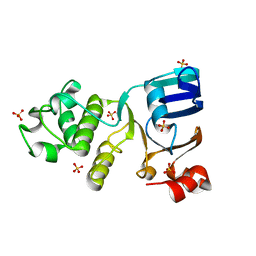 | | Nucleotide-free crystal structure of nucleotide-binding domain 1 from human MRP1 supports a general-base catalysis mechanism for ATP hydrolysis. | | Descriptor: | MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 1, SULFATE ION | | Authors: | Chaptal, V, Gueguen-Chaignon, V, Magnard, S, Falson, P, Di Pietro, A, Baubichon-Cortay, H. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nucleotide-Free Crystal Structure of Nucleotide-Binding Domain 1 from Human Abcc1 Supports a 'General-Base Catalysis' Mechanism for ATP Hydrolysis.
Biochem.Pharm., 3, 2014
|
|
4D5U
 
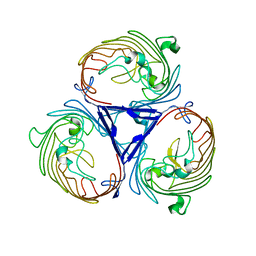 | | Structure of OmpF in I2 | | Descriptor: | OUTER MEMBRANE PROTEIN F | | Authors: | Chaptal, V, Kilburg, A, Flot, D, Wiseman, B, Aghajari, N, Jault, J.M, Falson, P. | | Deposit date: | 2014-11-07 | | Release date: | 2015-12-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Two Different Centered Monoclinic Crystals of the E. Coli Outer-Membrane Protein Ompf Originate from the Same Building Block.
Biochim.Biophys.Acta, 1858, 2016
|
|
4KSD
 
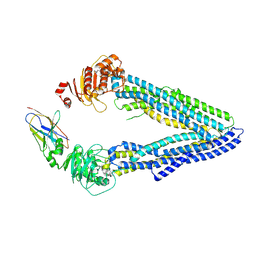 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A, R2 protein | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.1001 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KSC
 
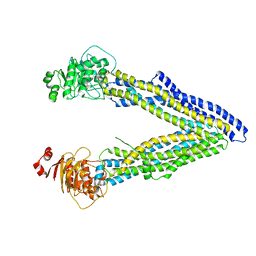 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KSB
 
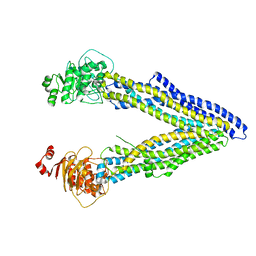 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8001 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JFB
 
 | | Crystal structure of OmpF in C2 with tNCS | | Descriptor: | Outer membrane protein F | | Authors: | Wiseman, B, Kilburg, A, Chaptal, V, Reyes-Meija, G.C, Sarwan, J, Falson, P, Jault, J.M. | | Deposit date: | 2013-02-28 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Stubborn contaminants: influence of detergents on the purity of the multidrug ABC transporter BmrA
PLoS ONE, 9, 2014
|
|
8RF1
 
 | | BmrA E504-R6G | | Descriptor: | Multidrug resistance ABC transporter ATP-binding/permease protein BmrA, RHODAMINE 6G | | Authors: | Gobet, A, Zarkadas, E, Schoehn, G, Falson, P, Chaptal, V. | | Deposit date: | 2023-12-12 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Rhodamine6G and Hœchst33342 narrow BmrA conformational spectrum for a more efficient use of ATP.
Nat Commun, 16, 2025
|
|
8REZ
 
 | | BmrA E504-apo | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Gobet, A, Zarkadas, E, Schoehn, G, Falson, P, Chaptal, V. | | Deposit date: | 2023-12-12 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Rhodamine6G and Hœchst33342 narrow BmrA conformational spectrum for a more efficient use of ATP.
Nat Commun, 16, 2025
|
|
8RGA
 
 | | BmrA E504-25uMATPMg | | Descriptor: | Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Gobet, A, Zarkadas, E, Schoehn, G, Falson, P, Chaptal, V. | | Deposit date: | 2023-12-13 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Rhodamine6G and Hœchst33342 narrow BmrA conformational spectrum for a more efficient use of ATP.
Nat Commun, 16, 2025
|
|
8RIA
 
 | | BmrA E504-100uMATPMg-IF | | Descriptor: | Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Gobet, A, Zarkadas, E, Schoehn, G, Falson, P, Chaptal, V. | | Deposit date: | 2023-12-18 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Rhodamine6G and Hœchst33342 narrow BmrA conformational spectrum for a more efficient use of ATP.
Nat Commun, 16, 2025
|
|
8RG7
 
 | | BmrA E504-R6G-25uMATP-Mg | | Descriptor: | Multidrug resistance ABC transporter ATP-binding/permease protein BmrA, RHODAMINE 6G | | Authors: | Gobet, A, Zarkadas, E, Schoehn, G, Falson, P, Chaptal, V. | | Deposit date: | 2023-12-13 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Rhodamine6G and Hœchst33342 narrow BmrA conformational spectrum for a more efficient use of ATP.
Nat Commun, 16, 2025
|
|
8RGN
 
 | | BmrA E504-R6G-70uMATPMg | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding/permease protein BmrA, ... | | Authors: | Gobet, A, Zarkadas, E, Schoehn, G, Falson, P, Chaptal, V. | | Deposit date: | 2023-12-14 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Rhodamine6G and Hœchst33342 narrow BmrA conformational spectrum for a more efficient use of ATP.
Nat Commun, 16, 2025
|
|
8RI1
 
 | | BmrA E504-100uMATPMg | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Gobet, A, Zarkadas, E, Schoehn, G, Falson, P, Chaptal, V. | | Deposit date: | 2023-12-18 | | Release date: | 2025-01-29 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rhodamine6G and Hœchst33342 narrow BmrA conformational spectrum for a more efficient use of ATP.
Nat Commun, 16, 2025
|
|
