1NBO
 
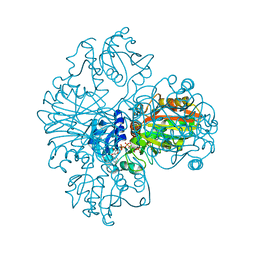 | | The dual coenzyme specificity of photosynthetic glyceraldehyde-3-phosphate dehydrogenase interpreted by the crystal structure of A4 isoform complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, glyceraldehyde-3-phosphate dehydrogenase A | | Authors: | Falini, G, Fermani, S, Ripamonti, A, Sabatino, P, Sparla, F, Pupillo, P, Trost, P. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dual Coenzyme Specificity of Photosynthetic Glyceraldehyde-3-phosphate
Dehydrogenase Interpreted by the Crystal Structure of A(4) Isoform
Complexed with NAD
Biochemistry, 42, 2003
|
|
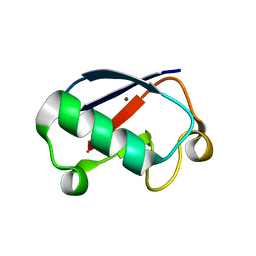
3EHV
 
 | |
3EFU
 
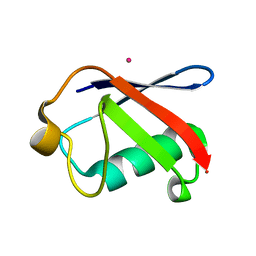 | |
3EEC
 
 | | X-ray structure of human ubiquitin Cd(II) adduct | | Descriptor: | CADMIUM ION, Ubiquitin | | Authors: | Falini, G, Fermani, S, Tosi, G, Arnesano, F, Natile, G. | | Deposit date: | 2008-09-04 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural probing of Zn(II), Cd(II) and Hg(II) binding to human ubiquitin.
Chem.Commun.(Camb.), 45, 2008
|
|
2PKR
 
 | | Crystal structure of (A+CTE)4 chimeric form of photosyntetic glyceraldehyde-3-phosphate dehydrogenase, complexed with NADP | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase Aor, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Fermani, S, Falini, G, Ripamonti, A. | | Deposit date: | 2007-04-18 | | Release date: | 2007-06-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6H7G
 
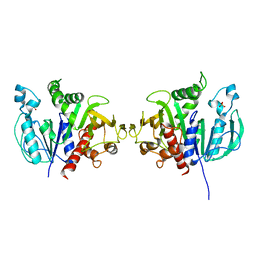 | | Crystal structure of redox-sensitive phosphoribulokinase (PRK) from the green algae Chlamydomonas reinhardtii | | Descriptor: | Phosphoribulokinase, chloroplastic, SULFATE ION | | Authors: | Fermani, S, Sparla, F, Gurrieri, L, Demitri, N, Polentarutti, M, Falini, G, Trost, P, Lemaire, S.D. | | Deposit date: | 2018-07-31 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ArabidopsisandChlamydomonasphosphoribulokinase crystal structures complete the redox structural proteome of the Calvin-Benson cycle.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2PKQ
 
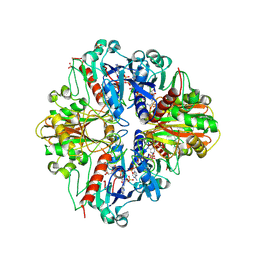 | | Crystal structure of the photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase, complexed with NADP | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, Glyceraldehyde-3-phosphate dehydrogenase B, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Fermani, S, Falini, G, Ripamonti, A. | | Deposit date: | 2007-04-18 | | Release date: | 2007-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2G5X
 
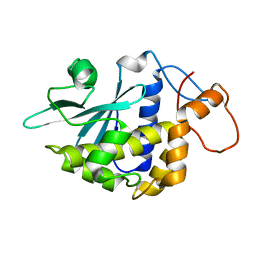 | | Crystal structure of lychnin a type 1 Ribosome Inactivating Protein (RIP) | | Descriptor: | Ribosome-inactivating protein | | Authors: | Fermani, S, Falini, G, Tosi, G, Ripamonti, A, Polito, L, Bolognesi, A, Stirpe, F. | | Deposit date: | 2006-02-23 | | Release date: | 2007-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of lychnin a type 1 Ribosome Inactivating Protein (RIP)
To be Published
|
|
8CO4
 
 | | Crystal structure of apo S-nitrosoglutathione reductase from Arabidopsis thalina | | Descriptor: | 1,2-ETHANEDIOL, Alcohol dehydrogenase class-3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fermani, S, Fanti, S, Carloni, G, Rossi, J, Falini, G, Zaffagnini, M. | | Deposit date: | 2023-02-27 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical characterization of Arabidopsis alcohol dehydrogenases reveals distinct functional properties but similar redox sensitivity.
Plant J., 118, 2024
|
|
8CON
 
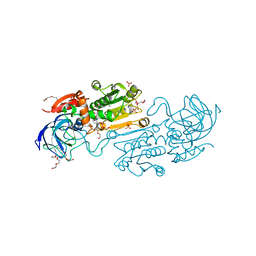 | | Crystal structure of alcohol dehydrogenase from Arabidopsis thaliana in complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase class-P, ... | | Authors: | Fermani, S, Fanti, S, Carloni, G, Falini, G, Meloni, M, Zaffagnini, M. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical characterization of Arabidopsis alcohol dehydrogenases reveals distinct functional properties but similar redox sensitivity.
Plant J., 118, 2024
|
|
6QUN
 
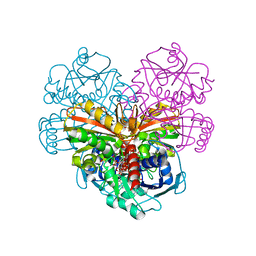 | | Crystal structure of AtGapC1 with the catalytic Cys149 irreversibly oxidized by H2O2 treatment | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase GAPC1, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Trost, P. | | Deposit date: | 2019-02-28 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Glutathionylation primes soluble glyceraldehyde-3-phosphate dehydrogenase for late collapse into insoluble aggregates.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6QUQ
 
 | | Crystal structure of glutathionylated glycolytic glyceraldehyde-3- phosphate dehydrogenase from Arabidopsis thaliana (AtGAPC1) | | Descriptor: | GLUTATHIONE, Glyceraldehyde-3-phosphate dehydrogenase GAPC1, cytosolic, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Trost, P. | | Deposit date: | 2019-02-28 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Glutathionylation primes soluble glyceraldehyde-3-phosphate dehydrogenase for late collapse into insoluble aggregates.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4K7U
 
 | | Crystal structure of Zn2.3-hUb (human ubiquitin) adduct from a solution 70 mM zinc acetate/1.3 mM hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fermani, S, Falini, G, Calvaresi, M, Bottoni, A, Arnesano, F, Natile, G. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Conformational selection of ubiquitin quaternary structures driven by zinc ions.
Chemistry, 19, 2013
|
|
4K7W
 
 | | Crystal structure of Zn3-hUb(human ubiquitin) adduct from a solution 100 mM zinc acetate/1.3 mM hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ZINC ION, ... | | Authors: | Fermani, S, Falini, G, Calvaresi, M, Bottoni, A, Arnesano, F, Natile, G. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Conformational selection of ubiquitin quaternary structures driven by zinc ions.
Chemistry, 19, 2013
|
|
4K7S
 
 | | Crystal structure of Zn2-hUb (human ubiquitin) adduct from a solution 35 mM zinc acetate/1.3 mM hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ZINC ION, ... | | Authors: | Fermani, S, Falini, G, Calvaresi, M, Bottoni, A, Arnesano, F, Natile, G. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Conformational selection of ubiquitin quaternary structures driven by zinc ions.
Chemistry, 19, 2013
|
|
4Z0H
 
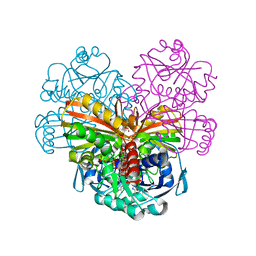 | | X-ray structure of cytoplasmic glyceraldehyde-3-phosphate dehydrogenase (GapC1) complexed with NAD | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase GAPC1, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Zaffagnini, M, Orru, R, Falini, G, Trost, P. | | Deposit date: | 2015-03-26 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tuning Cysteine Reactivity and Sulfenic Acid Stability by Protein Microenvironment in Glyceraldehyde-3-Phosphate Dehydrogenases of Arabidopsis thaliana.
Antioxid. Redox Signal., 24, 2016
|
|
3QV1
 
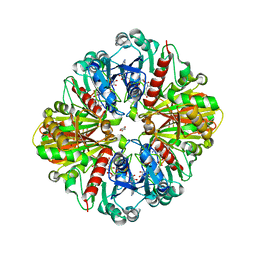 | | Crystal structure of the binary complex of photosyntetic A4 glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with cp12-2, both from Arabidopsis thaliana. | | Descriptor: | 1,2-ETHANEDIOL, CP12 protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Thumiger, A, Fermani, S, Falini, G, Marri, L, Sparla, F, Trost, P. | | Deposit date: | 2011-02-25 | | Release date: | 2012-02-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Selection and Folding-upon-binding of Intrinsically Disordered Protein CP12 Regulate Photosynthetic Enzymes Assembly.
J.Biol.Chem., 287, 2012
|
|
6H7H
 
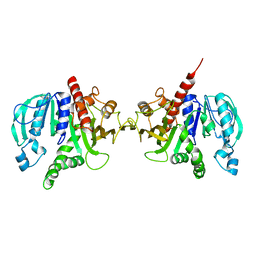 | | Crystal structure of redox-sensitive phosphoribulokinase (PRK) from Arabidopsis thaliana | | Descriptor: | Phosphoribulokinase, chloroplastic | | Authors: | Fermani, S, Sparla, F, Gurrieri, L, Falini, G, Trost, P. | | Deposit date: | 2018-07-31 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.471 Å) | | Cite: | ArabidopsisandChlamydomonasphosphoribulokinase crystal structures complete the redox structural proteome of the Calvin-Benson cycle.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3RVD
 
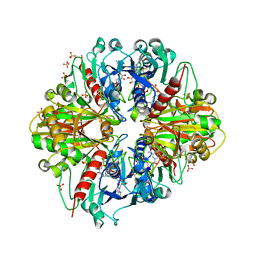 | | Crystal structure of the binary complex, obtained by soaking, of photosyntetic a4 glyceraldehyde 3-phosphate dehydrogenase (gapdh) with cp12-2, both from arabidopsis thaliana. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Thumiger, A, Falini, G, Marri, L, Sparla, F, Trost, P. | | Deposit date: | 2011-05-06 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational Selection and Folding-upon-binding of Intrinsically Disordered Protein CP12 Regulate Photosynthetic Enzymes Assembly.
J.Biol.Chem., 287, 2012
|
|
5NL4
 
 | | Crystal structure of Zn1.3-E16V human ubiquitin (hUb) mutant adduct, from a solution 35 mM zinc acetate/1.3 mM E16V hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
5NLI
 
 | | Crystal structure of Zn2-E16V human ubiquitin (hUb) mutant adduct, from a solution 35 mM zinc acetate/10% v/v TFE/1.3 mM E16V hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Polyubiquitin-C, ... | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
5NMC
 
 | |
5NL5
 
 | | Crystal structure of Zn1.7-E16V human ubiquitin (hUb) mutant adduct, from a solution 70 mM zinc acetate/1.3 mM E16V hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Polyubiquitin-B, ... | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
5NLF
 
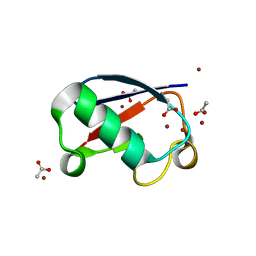 | | Crystal structure of Zn2.7-E16V human ubiquitin (hUb) mutant adduct, from a solution 100 mM zinc acetate/1.3 mM E16V hUb | | Descriptor: | ACETATE ION, Polyubiquitin-C, ZINC ION | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
5NLJ
 
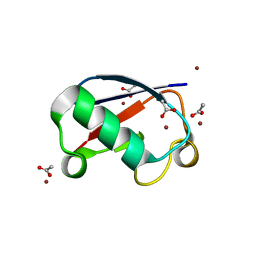 | | Crystal structure of Zn3-E16V human ubiquitin (hUb) mutant adduct, from a solution 70 mM zinc acetate/20% v/v TFE/1.3 mM E16V hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
