6QL4
 
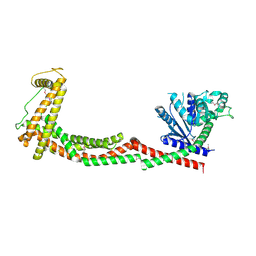 | | Crystal structure of nucleotide-free Mgm1 | | Descriptor: | 1,2-ETHANEDIOL, Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Wollweber, F, Pfitzner, A.-K, Muehleip, A, Sanchez, R, Kudryashev, M, Chiaruttin, N, Lilie, H, Schlegel, J, Rosenbaum, E, Hessenberger, M, Matthaeus, C, Noe, F, Roux, A, vanderLaan, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
3SNH
 
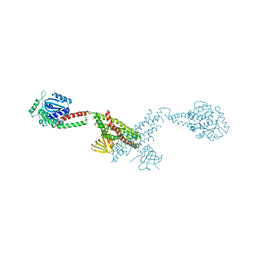 | |
6RZU
 
 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
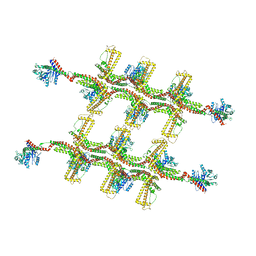
6RZV
 
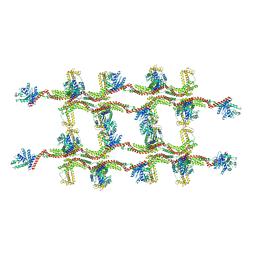 | | Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (20.6 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RZW
 
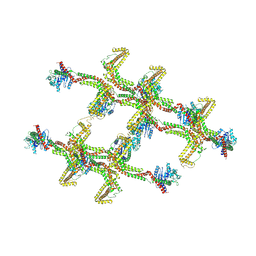 | | Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (18.799999 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RZT
 
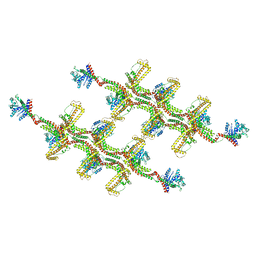 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
1JPS
 
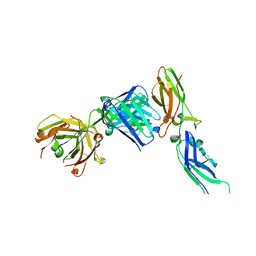 | | Crystal structure of tissue factor in complex with humanized Fab D3h44 | | Descriptor: | immunoglobulin Fab D3H44, heavy chain, light chain, ... | | Authors: | Faelber, K, Kirchhofer, D, Presta, L, Kelley, R.F, Muller, Y.A. | | Deposit date: | 2001-08-03 | | Release date: | 2002-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A resolution crystal structures of tissue factor in complex with humanized Fab D3h44 and of free humanized Fab D3h44: revisiting the solvation of antigen combining sites.
J.Mol.Biol., 313, 2001
|
|
1K6Q
 
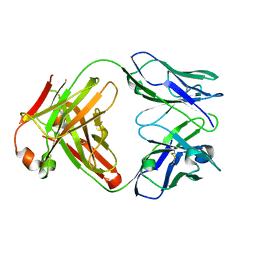 | | Crystal structure of antibody Fab fragment D3 | | Descriptor: | immunoglobulin Fab D3, heavy chain, light chain | | Authors: | Faelber, K, Kelley, R.F, Kirchhofer, D, Muller, Y.A. | | Deposit date: | 2001-10-17 | | Release date: | 2002-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of murine Fab D3 at 2.4 A resolution in comparison with the humanised Fab D3h44 (1.85A) provides structural insight into the humanisation process of the D3 anti-tissue factor antibody
To be Published
|
|
1JPT
 
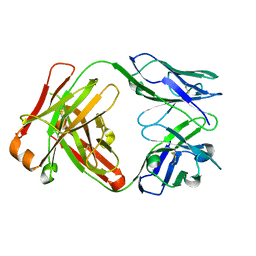 | | Crystal Structure of Fab D3H44 | | Descriptor: | immunoglobulin Fab D3H44, heavy chain, immunoglobulin Fab D3h44, ... | | Authors: | Faelber, K, Kirchhofer, D, Presta, L, Kelley, R.F, Muller, Y.A. | | Deposit date: | 2001-08-03 | | Release date: | 2002-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A resolution crystal structures of tissue factor in complex with humanized Fab D3h44 and of free humanized Fab D3h44: revisiting the solvation of antigen combining sites.
J.Mol.Biol., 313, 2001
|
|
3K3V
 
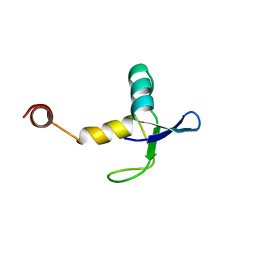 | |
3FMA
 
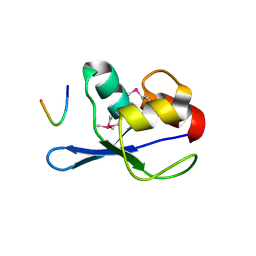 | |
4BEJ
 
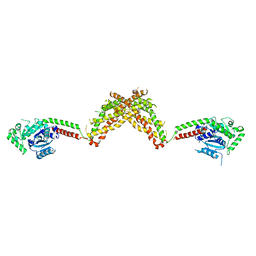 | | Nucleotide-free Dynamin 1-like protein (DNM1L, DRP1, DLP1) | | Descriptor: | DYNAMIN 1-LIKE PROTEIN | | Authors: | Froehlich, C, Schwefel, D, Faelber, K, Daumke, O. | | Deposit date: | 2013-03-11 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.483 Å) | | Cite: | Structural Insights Into Oligomerization and Mitochondrial Remodelling of Dynamin 1-Like Protein.
Embo J., 32, 2013
|
|
2NUZ
 
 | | crystal structure of alpha spectrin SH3 domain measured at room temperature | | Descriptor: | Spectrin alpha chain, brain | | Authors: | Agarwal, V, Faelber, K, Hologne, M, Chevelkov, V, Oschkinat, H, Diehl, A, Reif, B. | | Deposit date: | 2006-11-10 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparison of MAS solid-state NMR and X-ray crystallographic data in the analysis of protein dynamics in the solid state
To be Published
|
|
1U06
 
 | | crystal structure of chicken alpha-spectrin SH3 domain | | Descriptor: | AZIDE ION, Spectrin alpha chain, brain | | Authors: | Chevelkov, V, Faelber, K, Diehl, A, Heinemann, U, Oschkinat, H, Reif, B. | | Deposit date: | 2004-07-13 | | Release date: | 2005-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Detection of dynamic water molecules in a microcrystalline sample of the SH3 domain of alpha-spectrin by MAS solid-state NMR.
J.Biomol.Nmr, 31, 2005
|
|
5FPH
 
 | | The GTPase domains of the immunity-related Irga6 dimerize in a parallel head-to-head fashion | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, INTERFERON-INDUCIBLE GTPASE 1, MAGNESIUM ION, ... | | Authors: | Schulte, K, Pawlowski, N, Faelber, K, Froehlich, C, Howard, J, Daumke, O. | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Immunity-Related Gtpase Irga6 Dimerizes in a Parallel Head-to-Head Fashion.
Bmc Biol., 14, 2016
|
|
4ZP3
 
 | | AKAP18:PKA-RIIalpha structure reveals crucial anchor points for recognition of regulatory subunits of PKA | | Descriptor: | A-kinase anchor protein 7 isoforms alpha and beta, CADMIUM ION, cAMP-dependent protein kinase type II-alpha regulatory subunit | | Authors: | Goetz, F, Roske, Y, Faelber, K, Zuehlke, K, Autenrieth, K, Kreuchwig, A, Krause, G, Herberg, F.W, Daumke, O, Heinemann, U, Klussmann, E. | | Deposit date: | 2015-05-07 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | AKAP18:PKA-RII alpha structure reveals crucial anchor points for recognition of regulatory subunits of PKA.
Biochem.J., 473, 2016
|
|
5A3F
 
 | | Crystal structure of the dynamin tetramer | | Descriptor: | DYNAMIN 3 | | Authors: | Reubold, T.F, Faelber, K, Plattner, N, Posor, Y, Branz, K, Curth, U, Schlegel, J, Anand, R, Manstein, D.J, Noe, F, Haucke, V, Daumke, O, Eschenburg, S. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of the Dynamin Tetramer
Nature, 525, 2015
|
|
4YU0
 
 | | Crystal structure of a tetramer of GluA2 TR mutant ligand binding domains bound with glutamate at 1.26 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTAMIC ACID, Glutamate receptor 2,Glutamate receptor 2, ... | | Authors: | Chebli, M, Salazar, H, Baranovic, J, Carbone, A.L, Ghisi, V, Faelber, K, Lau, A.Y, Daumke, O, Plested, A.J.R. | | Deposit date: | 2015-03-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of the tetrameric GluA2 ligand-binding domain in complex with glutamate at 1.26 Angstroms resolution
To Be Published
|
|
4Z0I
 
 | | Crystal structure of a tetramer of GluA2 ligand binding domains bound with glutamate at 1.45 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTAMIC ACID, Glutamate receptor 2,Glutamate receptor 2, ... | | Authors: | Baranovic, J, Chebli, M, Salazar, H, Carbone, A.L, Ghisi, V, Faelber, K, Lau, A.Y, Daumke, O, Plested, A.J.R. | | Deposit date: | 2015-03-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the tetrameric wt GluA2 ligand-binding domain bound to glutamate at 1.45 Angstroms resolution
To Be Published
|
|
