1FDM
 
 | |
1ETU
 
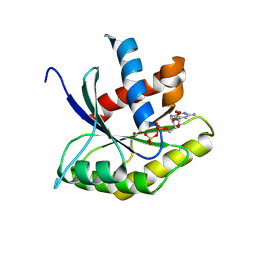 | | STRUCTURAL DETAILS OF THE BINDING OF GUANOSINE DIPHOSPHATE TO ELONGATION FACTOR TU FROM E. COLI AS STUDIED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | ELONGATION FACTOR TU, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Clark, B.F.C, Lacour, T.F.M, Kjeldgaard, M, Morikawa, K, Nyborg, J, Rubin, R, Thirup, S. | | Deposit date: | 1988-01-15 | | Release date: | 1988-07-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural details of the binding of guanosine diphosphate to elongation factor Tu from E. coli as studied by X-ray crystallography.
EMBO J., 4, 1985
|
|
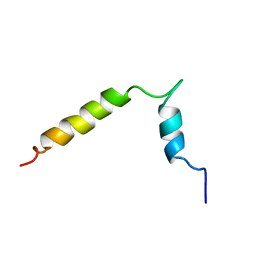
6FGM
 
 | | The NMR solution structure of the peptide AC12 from Hypsiboas raniceps | | Descriptor: | ALA-CYS-PHE-LEU-THR-ARG-LEU-GLY-THR-TYR-VAL-CYS | | Authors: | Popov, C.S.F.C, Simas, B.S, Goodfellow, B.J, Bocca, A.L, Andrade, P.B, Pereira, D, Valentao, P, Pereira, P.J.B, Rodrigues, J.E, Veloso Jr, P.H.H, Rezende, T.M.B. | | Deposit date: | 2018-01-11 | | Release date: | 2019-01-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Host-defense peptides AC12, DK16 and RC11 with immunomodulatory activity isolated from Hypsiboas raniceps skin secretion.
Peptides, 113, 2019
|
|
1JKZ
 
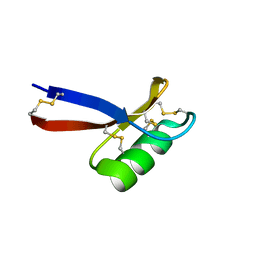 | | NMR Solution Structure of Pisum sativum defensin 1 (Psd1) | | Descriptor: | DEFENSE-RELATED PEPTIDE 1 | | Authors: | Almeida, M.S, Cabral, K.M.S, Kurtenbach, E, Almeida, F.C.L, Valente, A.P. | | Deposit date: | 2001-07-13 | | Release date: | 2002-02-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pisum sativum defensin 1 by high resolution NMR: plant defensins, identical backbone with different mechanisms of action.
J.Mol.Biol., 315, 2002
|
|
7N45
 
 | |
8TG0
 
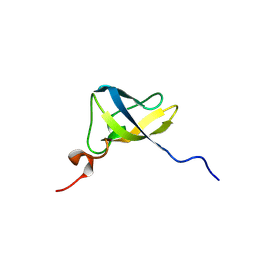 | |
6UI5
 
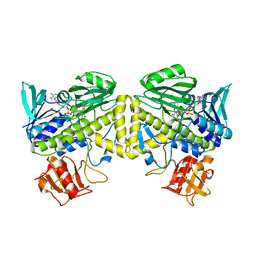 | |
6VPD
 
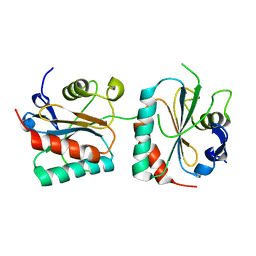 | | Crystal structure of Trgpx in apo form | | Descriptor: | Glutathione peroxidase | | Authors: | Adriani, P.P, De Oliveira, G.S, Paiva, F.C.R, Dias, M.V.B, Chambergo, F.S. | | Deposit date: | 2020-02-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural and functional characterization of the glutathione peroxidase-like thioredoxin peroxidase from the fungus Trichoderma reesei.
Int.J.Biol.Macromol., 167, 2020
|
|
6VK2
 
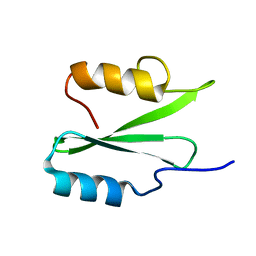 | |
6WUQ
 
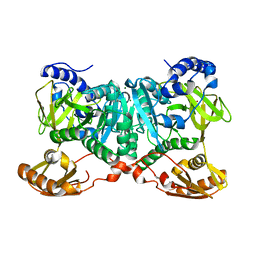 | | Crystal structure of AjiA1 in apo form | | Descriptor: | AjiA1, MAGNESIUM ION, ZINC ION | | Authors: | Paiva, F.C.R, Chan, K, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2020-05-05 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | The crystal structure of AjiA1 reveals a novel structural motion mechanism in the adenylate-forming enzyme family
Acta Crystallogr.,Sect.D, 76, 2020
|
|
5SZW
 
 | | NMR solution structure of the RRM1 domain of the post-transcriptional regulator HuR | | Descriptor: | ELAV-like protein 1 | | Authors: | Lixa, C, Mujo, A, Jendiroba, K.A, Almeida, F.C.L, Lima, L.M.T.R, Pinheiro, A.S. | | Deposit date: | 2016-08-15 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oligomeric transition and dynamics of RNA binding by the HuR RRM1 domain in solution.
J. Biomol. NMR, 72, 2018
|
|
6NK9
 
 | |
6NOI
 
 | | Crystal structure of Tsn15 in apo form | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Little, R, Paiva, F.C.R, Dias, M.V.B, Leadlay, P. | | Deposit date: | 2019-01-16 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unexpected enzyme-catalysed [4+2] cycloaddition and rearrangement in polyether antibiotic biosynthesis
Nat Catal, 2019
|
|
6NNW
 
 | | Tsn15 in complex with substrate intermediate | | Descriptor: | (3E)-3-{(1S,4S,4aS,5R,8aS)-1-[(2E,4R,7S,8E,10S)-1,7-dihydroxy-10-{(2R,3S,5R)-5-[(1S)-1-methoxyethyl]-3-methyloxolan-2-yl}-4-methylundeca-2,8-dien-2-yl]-4,5-dimethyloctahydro-3H-2-benzopyran-3-ylidene}oxolane-2,4-dione, PHOSPHATE ION, PROPANOIC ACID, ... | | Authors: | Paiva, F.C.R, Little, R, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2019-01-15 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unexpected enzyme-catalysed [4+2] cycloaddition and rearrangement in polyether antibiotic biosynthesis
Nat Catal, 2019
|
|
6NOM
 
 | | NMR solution structure of Pisum sativum defensin 2 (Psd2) provides evidence for the presence of hydrophobic surface clusters | | Descriptor: | Defensin-2 | | Authors: | Pinheiro-Aguiar, R, Amaral, V.S.G, Bastos, I, Kurtenbach, E, Almeida, F.C.L. | | Deposit date: | 2019-01-16 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of Pisum sativum defensin 2 provides evidence for the presence of hydrophobic surface-clusters.
Proteins, 88, 2020
|
|
6NAN
 
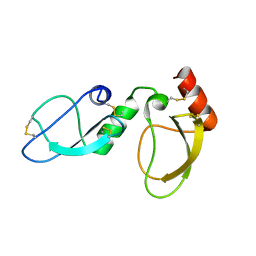 | | NMR structure determination of Ixolaris and Factor X interaction reveals a noncanonical mechanism of Kunitz inhibition | | Descriptor: | Ixolaris | | Authors: | De Paula, V.S, Sgourakis, N.G, Francischetti, I.M.B, Almeida, F.C.L, Monteiro, R.Q, Valente, A.P. | | Deposit date: | 2018-12-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of Ixolaris and factor X(a) interaction reveals a noncanonical mechanism of Kunitz inhibition.
Blood, 134, 2019
|
|
6C44
 
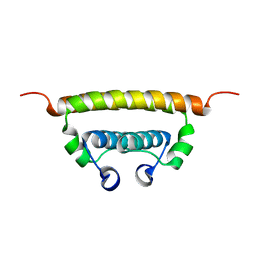 | | Zika virus capsid protein | | Descriptor: | Capsid protein | | Authors: | Morando, M.A, Barbosa, G.M, Cruz-Oliveira, C, Da Poian, A.T, Almeida, F.C.L. | | Deposit date: | 2018-01-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamics of Zika Virus Capsid Protein in Solution: The Properties and Exposure of the Hydrophobic Cleft Are Controlled by the alpha-Helix 1 Sequence.
Biochemistry, 58, 2019
|
|
6D6X
 
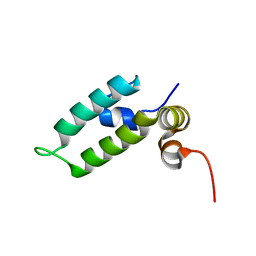 | | HSP40 co-chaperone Sis1 J-domain | | Descriptor: | Type II HSP40 co-chaperone | | Authors: | Pinheiro, G.M.S, Amorim, G.C, Iqbal, A, Ramos, C.H.I, Almeida, F.C.L. | | Deposit date: | 2018-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR investigation on the structure and function of the isolated J-domain from Sis1: Evidence of transient inter-domain interactions in the full-length protein.
Arch.Biochem.Biophys., 669, 2019
|
|
1S4H
 
 | | NMR structure of cross-reactive peptides from L. braziliensis | | Descriptor: | 60S acidic ribosomal protein P2 | | Authors: | Soares, M.R, Bisch, P.M, Campos de Carvalho, A.C, Valente, A.P, Almeida, F.C.L. | | Deposit date: | 2004-01-16 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Correlation between conformation and antibody binding: NMR structure of cross-reactive peptides from T. cruzi, human and L. braziliensis
Febs Lett., 560, 2004
|
|
1S4J
 
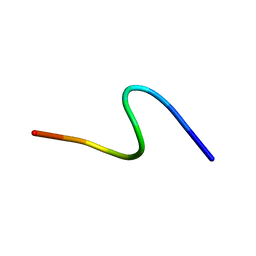 | | NMR structure of cross-reactive peptides from Homo sapiens | | Descriptor: | 60S acidic ribosomal protein P2 | | Authors: | Soares, M.R, Bisch, P.M, Campos de Carvalho, A.C, Valente, A.P, Almeida, F.C.L. | | Deposit date: | 2004-01-16 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Correlation between conformation and antibody binding: NMR structure of cross-reactive peptides from T. cruzi, human and L. braziliensis
Febs Lett., 560, 2004
|
|
1TTT
 
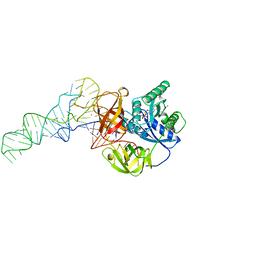 | | Phe-tRNA, elongation factoR EF-TU:GDPNP ternary complex | | Descriptor: | MAGNESIUM ION, OF ELONGATION FACTOR TU (EF-TU), PHENYLALANINE, ... | | Authors: | Nissen, P, Kjeldgaard, M, Thirup, S, Polekhina, G, Reshetnikova, L, Clark, B.F.C, Nyborg, J. | | Deposit date: | 1995-11-16 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog.
Science, 270, 1995
|
|
2I9H
 
 | |
2JX6
 
 | | Structure and membrane interactions of the antibiotic peptide dermadistinctin k by solution and oriented 15N and 31P solid-state NMR spectroscopy | | Descriptor: | Dermadistinctin-K | | Authors: | Mendonca Moraes, C, Verly, R.M, Resende, J.M, Bemquerer, M.P, Pilo-Veloso, D, Valente, A, Almeida, F.C.L, Bechinger, B. | | Deposit date: | 2007-11-08 | | Release date: | 2008-11-11 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy
Biophys.J., 96, 2009
|
|
2HSY
 
 | |
2JQ1
 
 | | Phylloseptin-3 | | Descriptor: | Phylloseptin-3 | | Authors: | Resende, J.M, Mendonca Moraes, C, Almeida, F.C.L, Prates, M.V, Cesar, A, Valente, A, Bemquerer, M.P, Pilo-Veloso, D, Bechinger, B. | | Deposit date: | 2007-05-25 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of the antimicrobial peptides phylloseptin-1, -2, and -3 and biological activity: The role of charges and hydrogen bonding interactions in stabilizing helix conformations
Peptides, 29, 2008
|
|
