2KVA
 
 | |
2L2K
 
 | |
2LSS
 
 | | Solution structure of the R. rickettsii cold shock-like protein | | Descriptor: | Cold shock-like protein | | Authors: | Veldkamp, C.T, Peterson, F.C, Gerarden, K.P, Fuchs, A.M, Koch, J.M, Mueller, M.M. | | Deposit date: | 2012-05-04 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cold-shock-like protein from Rickettsia rickettsii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2L47
 
 | |
2LC7
 
 | |
2L8R
 
 | |
2LQ7
 
 | |
2LGR
 
 | |
2L4N
 
 | |
2LC6
 
 | |
2LWL
 
 | | Structural Basis for the Interaction of Human β-Defensin 6 and Its Putative Chemokine Receptor CCR2 and Breast Cancer Microvesicles | | Descriptor: | Beta-defensin 106 | | Authors: | de Paula, V.S, Gomes, N.S.F, Lima, L.G, Miyamoto, C.A, Monteiro, R.Q, Almeida, F.C.L, Valente, A. | | Deposit date: | 2012-08-02 | | Release date: | 2013-08-21 | | Last modified: | 2013-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Interaction of Human beta-Defensin 6 and Its Putative Chemokine Receptor CCR2 and Breast Cancer Microvesicles.
J.Mol.Biol., 425, 2013
|
|
2M85
 
 | | PHD Domain from Human SHPRH | | Descriptor: | E3 ubiquitin-protein ligase SHPRH, ZINC ION | | Authors: | Machado, L.E.S.F, Pustovalova, Y, Pozhidaeva, A, Almeida, F.C.L, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2013-05-07 | | Release date: | 2013-08-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | PHD domain from human SHPRH.
J.Biomol.Nmr, 56, 2013
|
|
2LVX
 
 | |
2LWX
 
 | |
2MG4
 
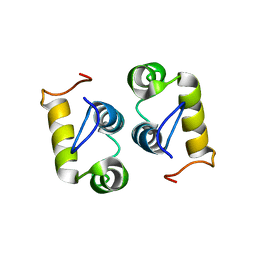 | | Computational design and experimental verification of a symmetric protein homodimer | | Descriptor: | Computational designed homodimer | | Authors: | Mou, Y, Huang, P.S, Hsu, F.C, Huang, S.J, Mayo, S.L. | | Deposit date: | 2013-10-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational design and experimental verification of a symmetric protein homodimer.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MBF
 
 | |
2MBX
 
 | | Structure, dynamics and stability of allergen cod parvalbumin Gad m 1 by solution and high-pressure NMR. | | Descriptor: | CALCIUM ION, Parvalbumin beta | | Authors: | Moraes, A.H, Ackerbauer, D, Bublin, M, Ferreira, F, Almeida, F.C.L, Breiteneder, H, Valente, A. | | Deposit date: | 2013-08-07 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution and high-pressure NMR studies of the structure, dynamics, and stability of the cross-reactive allergenic cod parvalbumin Gad m 1.
Proteins, 82, 2014
|
|
2MU4
 
 | | Structure of F. tularensis Virulence Determinant | | Descriptor: | flpp3Sol_2 | | Authors: | Zook, J.J.D.Z, Mo, G.G.M, Craciunescu, F.F.C, Sisco, N.N.S, Hansen, D.D.H, Baravati, B.B.B, Van Horn, W.W.V.H, Cherry, B.B.C, Fromme, P.P.F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Francisella tularensis Virulence Determinant Reveals Structural Homology to Bet v1 Allergen Proteins.
Structure, 23, 2015
|
|
2N5A
 
 | |
2MP1
 
 | |
2N1H
 
 | |
2N5B
 
 | |
2OJ8
 
 | | NMR structure of the UGUU tetraloop of Duck Epsilon apical stem loop of the Hepatitis B virus | | Descriptor: | 5'-R(P*GP*CP*UP*GP*UP*UP*GP*U)-3' | | Authors: | Girard, F.C, Ottink, O.M, Ampt, K.A.M, Tessari, M, Wijmenga, S.S. | | Deposit date: | 2007-01-12 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Thermodynamics and NMR studies on Duck, Heron and Human HBV encapsidation signals.
Nucleic Acids Res., 35, 2007
|
|
2OJ7
 
 | | NMR structure of the UGUU tetraloop of Duck Epsilon apical stem loop | | Descriptor: | 5'-R(P*GP*CP*UP*GP*UP*UP*GP*U)-3' | | Authors: | Girard, F.C, Ottink, O.M, Ampt, K.A.M, Tessari, M, Wijmenga, S.S. | | Deposit date: | 2007-01-12 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Thermodynamics and NMR studies on Duck, Heron and Human HBV encapsidation signals.
Nucleic Acids Res., 35, 2007
|
|
2N55
 
 | |
