5UR6
 
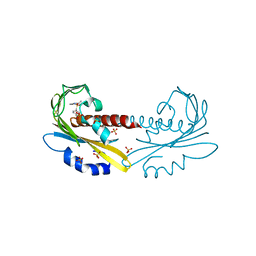 | | PYR1 bound to the rationally designed agonist cyanabactin | | Descriptor: | Abscisic acid receptor PYR1, N-(4-cyano-3-cyclopropylphenyl)-1-(4-methylphenyl)methanesulfonamide, SULFATE ION | | Authors: | Peterson, F.C, Vaidya, A, Jensen, D.R, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2017-02-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | A Rationally Designed Agonist Defines Subfamily IIIA Abscisic Acid Receptors As Critical Targets for Manipulating Transpiration.
ACS Chem. Biol., 12, 2017
|
|
5US5
 
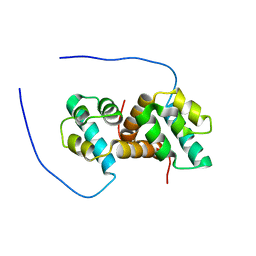 | | Solution structure of the IreB homodimer | | Descriptor: | UPF0297 protein EF_1202 | | Authors: | Lytle, B.L, Peterson, F.C, Volkman, B.F, Kristich, C.J, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2017-02-13 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dimerization of IreB, a Negative Regulator of Cephalosporin Resistance in Enterococcus faecalis.
J. Mol. Biol., 429, 2017
|
|
5UR4
 
 | |
5UR7
 
 | |
5SZW
 
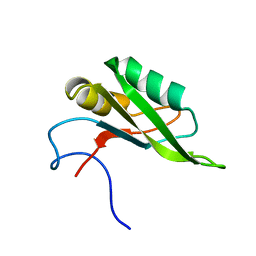 | | NMR solution structure of the RRM1 domain of the post-transcriptional regulator HuR | | Descriptor: | ELAV-like protein 1 | | Authors: | Lixa, C, Mujo, A, Jendiroba, K.A, Almeida, F.C.L, Lima, L.M.T.R, Pinheiro, A.S. | | Deposit date: | 2016-08-15 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oligomeric transition and dynamics of RNA binding by the HuR RRM1 domain in solution.
J. Biomol. NMR, 72, 2018
|
|
4OKJ
 
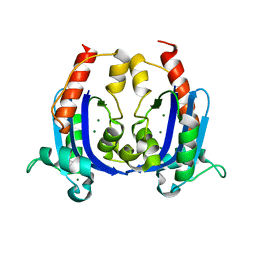 | | Crystal structure of RNase AS from M tuberculosis | | Descriptor: | 3'-5' exoribonuclease Rv2179c/MT2234.1, MAGNESIUM ION | | Authors: | Romano, M, van de Weerd, R, Brouwer, F.C.C, Roviello, G.N, Lacroix, R, Sparrius, M, van den Brink-van Stempvoort, G, Maaskant, J.J, van der Sar, A.M, Appelmelk, B.J, Geurtsen, J.J, Berisio, R. | | Deposit date: | 2014-01-22 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Function of RNase AS, a Polyadenylate-Specific Exoribonuclease Affecting Mycobacterial Virulence In Vivo
Structure, 22, 2014
|
|
4OKK
 
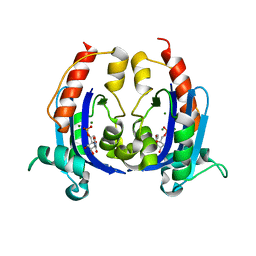 | | Crystal structure of RNase AS from M tuberculosis in complex with UMP | | Descriptor: | 3'-5' exoribonuclease Rv2179c/MT2234.1, MAGNESIUM ION, URIDINE-5'-MONOPHOSPHATE | | Authors: | Romano, M, van de Weerd, R, Brouwer, F.C.C, Roviello, G.N, Lacroix, R, Sparrius, M, van den Brink-van Stempvoort, G, Maaskant, J.J, van der Sar, A.M, Appelmelk, B.J, Geurtsen, J.J, Berisio, R. | | Deposit date: | 2014-01-22 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure and Function of RNase AS, a Polyadenylate-Specific Exoribonuclease Affecting Mycobacterial Virulence In Vivo
Structure, 22, 2014
|
|
4NDL
 
 | | Computational design and experimental verification of a symmetric homodimer | | Descriptor: | ENH-c2b, computational designed homodimer | | Authors: | Mou, Y, Huang, P.S, Hsu, F.C, Huang, S.J, Mayo, S.L. | | Deposit date: | 2013-10-26 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Computational design and experimental verification of a symmetric protein homodimer.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1JF7
 
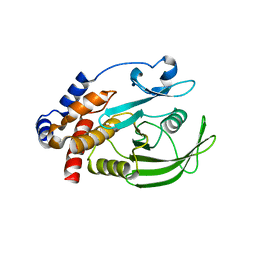 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXED WITH PNU177836 | | Descriptor: | 5-(2-{2-[(TERT-BUTOXY-HYDROXY-METHYL)-AMINO]-1-HYDROXY-3-PHENYL-PROPYLAMINO}-3-HYDROXY-3-PENTYLAMINO-PROPYL)-2-CARBOXYMETHOXY-BENZOIC ACID, PROTEIN-TYROSINE PHOSPHATASE 1B | | Authors: | Larsen, S.D, Barf, T, Liljebris, C, May, P.D, Ogg, D, O'Sullivan, T.J, Palazuk, B.J, Schostarez, H.J, Stevens, F.C, Bleasdale, J.E. | | Deposit date: | 2001-06-20 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and biological activity of a novel class of small molecular weight peptidomimetic competitive inhibitors of protein tyrosine phosphatase 1B.
J.Med.Chem., 45, 2002
|
|
6NK9
 
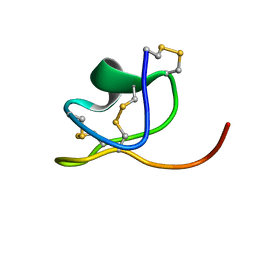 | |
1KQK
 
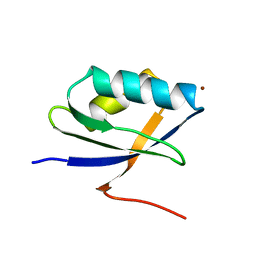 | | Solution Structure of the N-terminal Domain of a Potential Copper-translocating P-type ATPase from Bacillus subtilis in the Cu(I)loaded State | | Descriptor: | COPPER (I) ION, POTENTIAL COPPER-TRANSPORTING ATPASE | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, D'Onofrio, M, Gonnelli, L, Marhuenda-Egea, F.C, Ruiz-Duenas, F.J. | | Deposit date: | 2002-01-07 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis in the apo and Cu(I) loaded states.
J.Mol.Biol., 317, 2002
|
|
6NNW
 
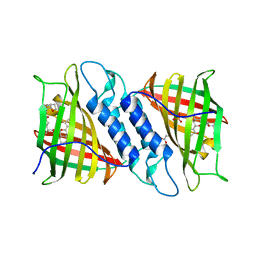 | | Tsn15 in complex with substrate intermediate | | Descriptor: | (3E)-3-{(1S,4S,4aS,5R,8aS)-1-[(2E,4R,7S,8E,10S)-1,7-dihydroxy-10-{(2R,3S,5R)-5-[(1S)-1-methoxyethyl]-3-methyloxolan-2-yl}-4-methylundeca-2,8-dien-2-yl]-4,5-dimethyloctahydro-3H-2-benzopyran-3-ylidene}oxolane-2,4-dione, PHOSPHATE ION, PROPANOIC ACID, ... | | Authors: | Paiva, F.C.R, Little, R, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2019-01-15 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unexpected enzyme-catalysed [4+2] cycloaddition and rearrangement in polyether antibiotic biosynthesis
Nat Catal, 2019
|
|
6NOI
 
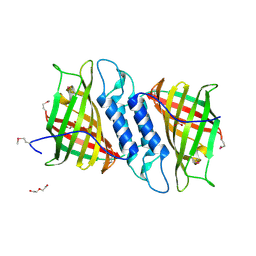 | | Crystal structure of Tsn15 in apo form | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Little, R, Paiva, F.C.R, Dias, M.V.B, Leadlay, P. | | Deposit date: | 2019-01-16 | | Release date: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unexpected enzyme-catalysed [4+2] cycloaddition and rearrangement in polyether antibiotic biosynthesis
Nat Catal, 2019
|
|
1JKZ
 
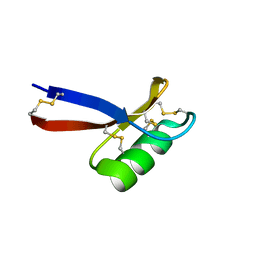 | | NMR Solution Structure of Pisum sativum defensin 1 (Psd1) | | Descriptor: | DEFENSE-RELATED PEPTIDE 1 | | Authors: | Almeida, M.S, Cabral, K.M.S, Kurtenbach, E, Almeida, F.C.L, Valente, A.P. | | Deposit date: | 2001-07-13 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pisum sativum defensin 1 by high resolution NMR: plant defensins, identical backbone with different mechanisms of action.
J.Mol.Biol., 315, 2002
|
|
6NWB
 
 | | PYL10 bound to the selective agonist hexabactin | | Descriptor: | Abscisic acid receptor PYL10, N-{[(4-cyanophenyl)methyl]sulfonyl}-1-(thiophen-3-yl)cyclohexane-1-carboxamide | | Authors: | Peterson, F.C, Vaidya, A, Jensen, D.R, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2019-02-06 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | PYL10 bound to the selective agonist hexabactin
To Be Published
|
|
6NWC
 
 | | PYL10 bound to the ABA pan-agonist 3CB | | Descriptor: | 1-{[(4-cyano-3-cyclopropylphenyl)acetyl]amino}cyclohexane-1-carboxylic acid, Abscisic acid receptor PYL10 | | Authors: | Peterson, F.C, Vaidya, A, Jensen, D.R, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2019-02-06 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Dynamic control of plant water use using designed ABA receptor agonists.
Science, 366, 2019
|
|
6NOM
 
 | | NMR solution structure of Pisum sativum defensin 2 (Psd2) provides evidence for the presence of hydrophobic surface clusters | | Descriptor: | Defensin-2 | | Authors: | Pinheiro-Aguiar, R, Amaral, V.S.G, Bastos, I, Kurtenbach, E, Almeida, F.C.L. | | Deposit date: | 2019-01-16 | | Release date: | 2019-08-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of Pisum sativum defensin 2 provides evidence for the presence of hydrophobic surface-clusters.
Proteins, 88, 2020
|
|
6NAN
 
 | | NMR structure determination of Ixolaris and Factor X interaction reveals a noncanonical mechanism of Kunitz inhibition | | Descriptor: | Ixolaris | | Authors: | De Paula, V.S, Sgourakis, N.G, Francischetti, I.M.B, Almeida, F.C.L, Monteiro, R.Q, Valente, A.P. | | Deposit date: | 2018-12-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of Ixolaris and factor X(a) interaction reveals a noncanonical mechanism of Kunitz inhibition.
Blood, 134, 2019
|
|
1MKE
 
 | | Structure of the N-WASP EVH1 Domain-WIP complex | | Descriptor: | Fusion protein consisting of Wiskott-Aldrich syndrome protein interacting protein (WIP), GSGSG linker, and Neural Wiskott-Aldrich syndrome protein (N-WASP) | | Authors: | Volkman, B.F, Prehoda, K.E, Scott, J.A, Peterson, F.C, Lim, W.A. | | Deposit date: | 2002-08-29 | | Release date: | 2002-12-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-WASP EVH1 Domain-WIP Complex. Insight into the
Molecular Basis of Wiskott-Aldrich Syndrome.
Cell(Cambridge,Mass.), 111, 2002
|
|
1M5A
 
 | |
1JMJ
 
 | | Crystal Structure of Native Heparin Cofactor II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HEPARIN COFACTOR II, ... | | Authors: | Baglin, T.P, Carrell, R.W, Church, F.C, Huntington, J.A. | | Deposit date: | 2001-07-18 | | Release date: | 2002-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of native and thrombin-complexed heparin cofactor II reveal a multistep allosteric mechanism.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1M02
 
 | | NMR Structure of PW2 Bound to SDS Micelles: A Tryptophan-rich Anticocidial Peptide Selected from Phage Display Libraries | | Descriptor: | HIS-PRO-LEU-LYS-GLN-TYR-TRP-TRP-ARG-PRO-SER-ILE | | Authors: | Tinoco, L.W, da Silva Jr, A, Leite, A, Valente, A.P, Almeida, F.C. | | Deposit date: | 2002-06-11 | | Release date: | 2002-08-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of PW2 bound to SDS micelles. A tryptophan-rich anticoccidial peptide selected from phage display libraries
J.Biol.Chem., 277, 2002
|
|
6ALK
 
 | | NMR solution structure of the major beech pollen allergen Fag s 1 | | Descriptor: | Fag s 1 pollen allergen | | Authors: | Moraes, A.H, Asam, A, Almeida, F.C.L, Wallner, M, Ferreira, F, Valente, A.P. | | Deposit date: | 2017-08-08 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for cross-reactivity and conformation fluctuation of the major beech pollen allergen Fag s 1.
Sci Rep, 8, 2018
|
|
6AX2
 
 | |
6B9W
 
 | |
