4DNM
 
 | | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound hepes, space group p3221 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Amidohydrolase 2, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-08 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound hepes, space group p3221
to be published
|
|
4DEJ
 
 | | Crystal structure of glutathione transferase-like protein IL0419 (Target EFI-501089) from Idiomarina loihiensis L2TR | | Descriptor: | Glutathione S-transferase related protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-20 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase-Like Protein Il0419 from Idiomarina Loihiensis
To be Published
|
|
4KH7
 
 | | Crystal structure of a glutathione transferase family member from salmonella enterica ty2, target efi-507262, with bound glutathione | | Descriptor: | GLUTATHIONE, Glutathione s-transferase family protein | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-30 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Salmonella enterica ty2, target efi-507262, with bound glutathione
To be published
|
|
4EEK
 
 | | Crystal structure of HAD FAMILY HYDROLASE DR_1622 from Deinococcus radiodurans R1 (TARGET EFI-501256) With bound phosphate and sodium | | Descriptor: | Beta-phosphoglucomutase-related protein, PHOSPHATE ION, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of HAD HYDROLASE DR_1622 Deinococcus radiodurans R1 (TARGET EFI-501256)
To be Published
|
|
4EEL
 
 | | Crystal structure of HAD FAMILY HYDROLASE DR_1622 from Deinococcus radiodurans R1 (TARGET EFI-501256) with bound citrate and sodium | | Descriptor: | Beta-phosphoglucomutase-related protein, CITRIC ACID, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of HAD HYDROLASE DR_1622 Deinococcus radiodurans R1 (TARGET EFI-501256)
To be Published
|
|
4EEN
 
 | | crystal structure of HAD FAMILY HYDROLASE DR_1622 from Deinococcus radiodurans R1 (TARGET EFI-501256) with bound magnesium | | Descriptor: | Beta-phosphoglucomutase-related protein, CHLORIDE ION, MAGNESIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of HAD HYDROLASE DR_1622 Deinococcus radiodurans R1 (TARGET EFI-501256)
To be Published
|
|
4F4R
 
 | | Crystal structure of D-mannonate dehydratase homolog from Chromohalobacter salexigens (Target EFI-502114), with bound NA, ordered loop | | Descriptor: | CHLORIDE ION, D-mannonate dehydratase, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2012-05-11 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of D-mannonate dehydratase homolog from Chromohalobacter salexigens (Target EFI-502114), with bound NA, ordered loop
to be published
|
|
4KGI
 
 | | Crystal structure of a glutathione transferase family member from Shigella flexneri, target EFI-507258, bound GSH, TEV-his-tag linker in active site | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Shigella flexneri, target EFI-507258, bound GSH, TEV-His-tag linker in active site
To be published
|
|
4GME
 
 | | Crystal structure of mannonate dehydratase (target EFI-502209) from caulobacter crescentus cb15 complexed with magnesium and d-mannonate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-MANNONIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-15 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mannonate Dehydratase from Caulobacter Crescentus Cb15
To be Published
|
|
4GP1
 
 | | Crystal structure of ISOPRENOID SYNTHASE A3MSH1 (TARGET EFI-501992) from pyrobaculum calidifontis complexed with DMAPP | | Descriptor: | CHLORIDE ION, DIMETHYLALLYL DIPHOSPHATE, Polyprenyl synthetase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Isoprenoid Synthase from Pyrobaculum Calidifontis
To be Published
|
|
4MP4
 
 | | Crystal structure of a glutathione transferase family member from Acinetobacter baumannii, Target EFI-501785, apo structure | | Descriptor: | Glutathione S-transferase, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Acinetobacter baumannii, Target EFI-501785, apo structure
To be Published
|
|
4GRF
 
 | | Crystal structure of thioredoxin domain of thiol-disulfide oxidoreductase BVU-2223 (Target EFI-501010) from Bacteroides vulgatus | | Descriptor: | CHLORIDE ION, GLUTATHIONE, Putative thiol-disulfide oxidoreductase, ... | | Authors: | Patskovsky, Y, Sampathkumar, P, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Imker, H.J, Hammonds, J, Al-Obaidi, N.F, Stead, M, Love, J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-24 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of thioredoxin domain of thiol-disulfide oxidoreductase BVU-2223 from Bacteroides vulgatus
To be Published
|
|
3TOU
 
 | | Crystal structure of GLUTATHIONE TRANSFERASE (TARGET EFI-501058) from Ralstonia solanacearum GMI1000 with GSH bound | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione s-transferase protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-06 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of GLUTATHIONE S-TRANSFERASE from Ralstonia solanacearum
To be Published
|
|
3UBK
 
 | | Crystal structure of glutathione transferase (TARGET EFI-501770) from leptospira interrogans | | Descriptor: | CHLORIDE ION, GLYCEROL, Glutathione transferase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-24 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Leptospira Interrogans
To be Published
|
|
3SJN
 
 | | Crystal structure of enolase Spea_3858 (target EFI-500646) from Shewanella pealeana with magnesium bound | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein, ... | | Authors: | Patskovsky, Y, Kim, J, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Enolase Spea_3858 from Shewanella Pealeana
To be Published
|
|
4QRZ
 
 | | Crystal structure of sugar transporter atu4361 from agrobacterium fabrum c58, target efi-510558, with bound maltotriose | | Descriptor: | ABC-TYPE SUGAR TRANSPORTER, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Love, J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-07-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of Maltoside Transporter from Agrobacterium Radiobacter, Target Efi-510558
To be Published
|
|
4HOJ
 
 | | Crystal structure of glutathione transferase homolog from Neisseria Gonorrhoeae, target EFI-501841, with bound glutathione | | Descriptor: | ACETATE ION, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-10-22 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of glutathione transferase homolog from Neisseria Gonorrhoeae, target EFI-501841, with bound glutathione
To be Published
|
|
4JB3
 
 | | Crystal structure of BT_0970, a had family phosphatase from bacteroides thetaiotaomicron VPI-5482, TARGET EFI-501083, with bound sodium and glycerol, closed lid, ordered loop | | Descriptor: | GLYCEROL, Haloacid dehalogenase-like hydrolase, SODIUM ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Kumar, P.R, Ghosh, A, Al Obaidi, N.F, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of bt_0970, a had family phosphatase from bacteroides thetaiotaomicron VPI-5482, TARGET EFI-501083, with bound sodium and glycerol, closed lid, ordered loop
To be Published
|
|
4JBD
 
 | | Crystal structure of Pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group I2, bound citrate | | Descriptor: | CITRIC ACID, Proline racemase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of pput_1285, a putative hydroxyproline epimerase from pseudomonas putida f1 (target efi-506500), open form, space group i2, bound citrate
To be Published
|
|
4JKU
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with quinaldic acid, NYSGRC Target 14306 | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, Probable sugar kinase protein, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-11 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with quinaldic acid, NYSGRC Target 14306
To be Published
|
|
4JD7
 
 | | Crystal structure of pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group P212121, bound sulfate | | Descriptor: | Proline racemase, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-24 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group P212121, bound sulfate
To be Published
|
|
4JN8
 
 | | Crystal structure of an enolase (putative galactarate dehydratase, target efi-500740) from agrobacterium radiobacter, bound sulfate, no metal ion, ordered active site | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ENOLASE, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Bouvier, J.T, Wichelecki, D, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an enolase (putative galactarate dehydratase, target efi-500740) from agrobacterium radiobacter, bound sulfate, no metal ion, ordered active site
To be Published
|
|
4JN7
 
 | | CRYSTAL STRUCTURE OF AN ENOLASE (PUTATIVE GALACTARATE DEHYDRATASE, TARGET EFI-500740) FROM AGROBACTERIUM RADIOBACTER, BOUND NA and L-MALATE, ORDERED ACTIVE SITE | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Bouvier, J.T, Wichelecki, D, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ENOLASE (PUTATIVE GALACTARATE DEHYDRATASE, TARGET EFI-500740) FROM AGROBACTERIUM RADIOBACTER, BOUND NA and L-MALATE, ORDERED ACTIVE SITE
To be Published
|
|
4K9C
 
 | | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N-(HYDROXYMETHYL)BENZAMIDE and 4-METHYL-3,4-DIHYDRO-2H-1,4-BENZOXAZINE-7-CARBOXYLIC ACID | | Descriptor: | 4-methyl-3,4-dihydro-2H-1,4-benzoxazine-7-carboxylic acid, ADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N-(HYDROXYMETHYL)BENZAMIDE and 4-METHYL-3,4-DIHYDRO-2H-1,4-BENZOXAZINE-7-CARBOXYLIC ACID
To be Published
|
|
4KF9
 
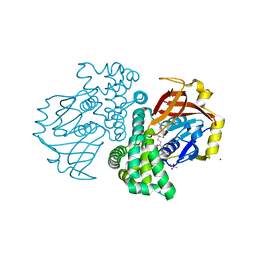 | | Crystal structure of a glutathione transferase family member from ralstonia solanacearum, target efi-501780, with bound gsh coordinated to a zinc ion, ordered active site | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione s-transferase protein, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-26 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a glutathione transferase family member from ralstonia solanacearum, target efi-501780, with bound gsh coordinated to a zinc ion, ordered active site
To be Published
|
|
