5M22
 
 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 | | Descriptor: | FE (III) ION, Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit | | Authors: | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | Deposit date: | 2016-10-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
5M4O
 
 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 in complex with 4-nitrophenol | | Descriptor: | FE (III) ION, Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit, ... | | Authors: | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | Deposit date: | 2016-10-18 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
5LJT
 
 | | Crystal structure of human carbonic anhydrase II in complex with the 4-((1-phenyl-1H-1,2,3-triazol-4-yl)methoxy)benzenesulfonamide inhibitor | | Descriptor: | 4-[[4-[azanyl-bis(oxidanyl)-$l^{4}-sulfanyl]phenoxy]methyl]-1-phenyl-1,2,3-triazole, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C. | | Deposit date: | 2016-07-19 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Benzenesulfonamides Incorporating Flexible Triazole Moieties Are Highly Effective Carbonic Anhydrase Inhibitors: Synthesis and Kinetic, Crystallographic, Computational, and Intraocular Pressure Lowering Investigations.
J. Med. Chem., 59, 2016
|
|
6H3Q
 
 | | Crystal structure of human carbonic anhydrase II in complex with the 4-(5-(chloromethyl)-1,3-selenazol-2-yl)benzenesulfonamide | | Descriptor: | 4-[5-(chloromethyl)-1,3-selenazol-2-yl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Ferraroni, M, Angeli, A, Supuran, C. | | Deposit date: | 2018-07-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Discovery of new 2, 5-disubstituted 1,3-selenazoles as selective human carbonic anhydrase IX inhibitors with potent anti-tumor activity.
Eur.J.Med.Chem., 157, 2018
|
|
6G3V
 
 | | Crystal structure of human carbonic anhydrase I in complex with the inhibitor famotidine | | Descriptor: | Carbonic anhydrase 1, GLYCEROL, ZINC ION, ... | | Authors: | Ferraroni, M, Supuran, C.T, Angeli, A. | | Deposit date: | 2018-03-26 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Famotidine, an Antiulcer Agent, Strongly InhibitsHelicobacter pyloriand Human Carbonic Anhydrases.
ACS Med Chem Lett, 9, 2018
|
|
4P1D
 
 | | Structure of the complex of a bimolecular human telomeric DNA with Coptisine | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, POTASSIUM ION, telomeric DNA | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P. | | Deposit date: | 2014-02-26 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the complex of a bimolecular human telomeric DNA with Coptisine
to be published
|
|
1MFM
 
 | | MONOMERIC HUMAN SOD MUTANT F50E/G51E/E133Q AT ATOMIC RESOLUTION | | Descriptor: | CADMIUM ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Rypniewski, W, Wilson, K.S, Orioli, P.L, Viezzoli, M.S, Banci, L, Bertini, I, Mangani, S. | | Deposit date: | 1999-04-16 | | Release date: | 1999-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The crystal structure of the monomeric human SOD mutant F50E/G51E/E133Q at atomic resolution. The enzyme mechanism revisited.
J.Mol.Biol., 288, 1999
|
|
3NW4
 
 | | Crystal Structure of Salicylate 1,2-dioxygenase G106A mutant from Pseudoaminobacter salicylatoxidans in complex with gentisate | | Descriptor: | 2,5-dihydroxybenzoic acid, FE (II) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-07-09 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The salicylate 1,2-dioxygenase as a model for a conventional gentisate 1,2-dioxygenase: crystal structures of the G106A mutant and its adducts with gentisate and salicylate.
FEBS J., 280, 2013
|
|
1SXS
 
 | | Reduced bovine superoxide dismutase at pH 5.0 complexed with thiocyanate | | Descriptor: | CALCIUM ION, COPPER (II) ION, PROTEIN (CU-ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Ferraroni, M, Rypniewski, W.R, Bruni, B, Orioli, P, Mangani, S. | | Deposit date: | 1998-09-24 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic determination of reduced bovine superoxide dismutase at pH 5.0 and of anion binding to its active site
J.Biol.Inorg.Chem., 3, 1998
|
|
1SXZ
 
 | | Reduced bovine superoxide dismutase at pH 5.0 complexed with azide | | Descriptor: | AZIDE ION, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Rypniewski, W.R, Bruni, B, Orioli, P, Wilson, K.S, Mangani, S. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic determination of reduced bovine superoxide dismutase at pH 5.0 and of anion binding to its active site
J.Biol.Inorg.Chem., 3, 1998
|
|
5N25
 
 | | Crystal structure of human carbonic anhydrase II in complex with the inhibitor 4-Pyridin-3-yl-benzenesulfonamide | | Descriptor: | 4-pyridin-3-ylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T, Scozzafava, A, Carta, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Sulfonamide carbonic anhydrase inhibitors: Zinc coordination and tail effects influence inhibitory efficacy and selectivity for different isoforms
Inorg.Chim.Acta., 470, 2018
|
|
5N1S
 
 | | Crystal structure of human carbonic anhydrase II in complex with the inhibitor 4-(1H-Indol-2-yl)-benzenesulfonamide | | Descriptor: | 4-(1~{H}-indol-2-yl)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T, Scozzafava, A, Carta, F. | | Deposit date: | 2017-02-06 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Sulfonamide carbonic anhydrase inhibitors: Zinc coordination and tail effects influence inhibitory efficacy and selectivity for different isoforms
Inorg.Chim.Acta., 470, 2018
|
|
5N24
 
 | | Crystal structure of human carbonic anhydrase II in complex with the inhibitor b4'-Cyano-biphenyl-4-sulfonic acid amide | | Descriptor: | 4-(4-cyanophenyl)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T, Scozzafava, A, Carta, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sulfonamide carbonic anhydrase inhibitors: Zinc coordination and tail effects influence inhibitory efficacy and selectivity for different isoforms
Inorg.Chim.Acta., 470, 2018
|
|
4L5K
 
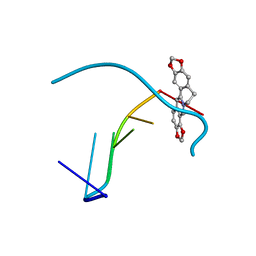 | | Crystal structure of the complex of DNA hexamer d(CGATCG) with Coptisine | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P. | | Deposit date: | 2013-06-11 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of the complex of DNA hexamer d(CGATCG) with Coptisine
to be published
|
|
3R6R
 
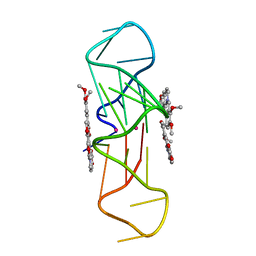 | | Structure of the complex of an intramolecular human telomeric DNA with Berberine formed in K+ solution | | Descriptor: | BERBERINE, DNA (22-mer), POTASSIUM ION | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Bilia, A.R. | | Deposit date: | 2011-03-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of human telomeric DNA complexed with berberine: an interesting case of stacked ligand to G-tetrad ratio higher than 1:1.
Nucleic Acids Res., 41, 2013
|
|
5N1R
 
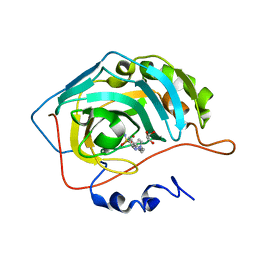 | | Crystal structure of human carbonic anhydrase II in complex with the inhibitor 4'-Pyrazol-1-ylmethyl-biphenyl-4-sulfonamide | | Descriptor: | 4-[4-(pyrazol-1-ylmethyl)phenyl]benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T, Scozzafava, A, Carta, F. | | Deposit date: | 2017-02-06 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Sulfonamide carbonic anhydrase inhibitors: Zinc coordination and tail effects influence inhibitory efficacy and selectivity for different isoforms
Inorg.Chim.Acta., 470, 2018
|
|
3NJZ
 
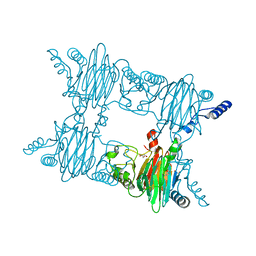 | | Crystal Structure of Salicylate 1,2-dioxygenase from Pseudoaminobacter salicylatoxidans Adducts with salicylate | | Descriptor: | 2-HYDROXYBENZOIC ACID, FE (II) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-06-18 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of salicylate 1,2-dioxygenase-substrates adducts: A step towards the comprehension of the structural basis for substrate selection in class III ring cleaving dioxygenases.
J.Struct.Biol., 177, 2012
|
|
3NL1
 
 | | Crystal Structure of Salicylate 1,2-dioxygenase from Pseudoaminobacter salicylatoxidans Adducts with gentisate | | Descriptor: | 2,5-dihydroxybenzoic acid, FE (II) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Crystal structures of salicylate 1,2-dioxygenase-substrates adducts: A step towards the comprehension of the structural basis for substrate selection in class III ring cleaving dioxygenases.
J.Struct.Biol., 177, 2012
|
|
3NKT
 
 | | Crystal Structure of Salicylate 1,2-dioxygenase from Pseudoaminobacter salicylatoxidans Adducts with naphthoate | | Descriptor: | 1-hydroxynaphthalene-2-carboxylic acid, FE (II) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of salicylate 1,2-dioxygenase-substrates adducts: A step towards the comprehension of the structural basis for substrate selection in class III ring cleaving dioxygenases.
J.Struct.Biol., 177, 2012
|
|
3NP6
 
 | | The crystal structure of Berberine bound to DNA d(CGTACG) | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', BERBERINE, CALCIUM ION | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Bilia, A.R. | | Deposit date: | 2010-06-28 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray diffraction analyses of the natural isoquinoline alkaloids Berberine and Sanguinarine complexed with double helix DNA d(CGTACG)
Chem.Commun.(Camb.), 47, 2011
|
|
3NX5
 
 | | The crystal structure of Sanguinarine bound to DNA d(CGTACG) | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, 5'-D(*CP*GP*TP*AP*CP*G)-3', CALCIUM ION | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Bilia, A.R. | | Deposit date: | 2010-07-13 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | X-Ray diffraction analyses of the natural isoquinoline alkaloids Berberine and Sanguinarine complexed with double helix DNA d(CGTACG)
Chem.Commun.(Camb.), 47, 2011
|
|
3NST
 
 | | Crystal Structure of Salicylate 1,2-dioxygenase G106A mutant from Pseudoaminobacter salicylatoxidans | | Descriptor: | FE (II) ION, GLYCEROL, Gentisate 1,2-dioxygenase | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-07-02 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The salicylate 1,2-dioxygenase as a model for a conventional gentisate 1,2-dioxygenase: crystal structures of the G106A mutant and its adducts with gentisate and salicylate.
FEBS J., 280, 2013
|
|
3NVC
 
 | | Crystal Structure of Salicylate 1,2-dioxygenase G106A mutant from Pseudoaminobacter salicylatoxidans in complex with salicylate | | Descriptor: | 2-HYDROXYBENZOIC ACID, FE (II) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-07-08 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The salicylate 1,2-dioxygenase as a model for a conventional gentisate 1,2-dioxygenase: crystal structures of the G106A mutant and its adducts with gentisate and salicylate.
FEBS J., 280, 2013
|
|
1SXN
 
 | | REDUCED BOVINE SUPEROXIDE DISMUTASE AT PH 5.0 | | Descriptor: | CALCIUM ION, COPPER (II) ION, CU, ... | | Authors: | Ferraroni, M, Rypniewski, W.R, Bruni, B, Orioli, P, Wilson, K.S, Mangani, S. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic determination of reduced bovine superoxide dismutase at pH 5.0 and of anion binding to its active site.
J.Biol.Inorg.Chem., 3, 1998
|
|
1M45
 
 | |
